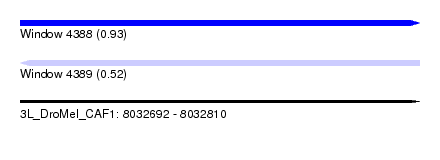
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,032,692 – 8,032,810 |
| Length | 118 |
| Max. P | 0.934200 |

| Location | 8,032,692 – 8,032,810 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -47.14 |
| Consensus MFE | -37.70 |
| Energy contribution | -39.70 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8032692 118 + 23771897 GCGCGAAGCUCGGUUUGGAGAGCGUCU--CUCCAGCUUUGUCGAGCUUUGUGCCCAUGCGUGCGAGAUCUUCGCCGGAAAGCUCUGACAAAUUCGGAAUCAGGCUGAGAUUUCAAUCGCG (((((((((((((.((((((((...))--)))))).....)))))))))))))....(((...((((((((.(((.((....(((((.....))))).)).))).))))))))...))). ( -52.70) >DroSec_CAF1 4142 118 + 1 GCGCGAAGCUCGGUUUGGAGAGCGUCG--CUCCAGCUUUGUCGAGCUUUGUGCCCAUGCGUGCGAGAUCUUCGCCGGAAAGCUCUGACCAAUUCGAAAUCAGGCUGAGAUUUCAAUCGCG (((((((((((((.((((((.......--)))))).....)))))))))))))....(((...((((((((.((((((....)))((.....)).......))).))))))))...))). ( -45.00) >DroSim_CAF1 4037 118 + 1 GCGCGAAGCUCGGUUUGGAGAGCGUCU--CUCCAGCUUUGUCGAGCUUUGUGCCCAUGCGUGCGAGAUCUUCGCCGGAAAGCUCUGACCAAUUCGAAAUCAGGCUGAGAUUUCAAUCGCG (((((((((((((.((((((((...))--)))))).....)))))))))))))....(((...((((((((.((((((....)))((.....)).......))).))))))))...))). ( -48.40) >DroEre_CAF1 4093 120 + 1 GCGCGAAGCUCGGUUUGGAGAGCGUCUCUCUCCAGCUUUGGCGAGCUUUGUGCCCAUGCGUACGAGAUCUUCACCGGAAAGCUCUGACCCAUUCGGAAUCAGGCUGAGCGUUCAAUCGAG ((((((((((((.(((((((((.....))))))))....).))))))))))))..((((........(((.....))).(((.((((.((....))..)))))))..))))......... ( -41.60) >DroYak_CAF1 4154 118 + 1 GCGCGAAGCUCGGUUUGGAGAGCGUCU--CUCCAGCUUUGGCGAGCUUUAUGCCCAUGCGUAUGAGAUCUUCGCCGGAAAGCUCUGACCCAUUGGGAGUCAGGCUGAGCUUUCAAUCGAG ....((((((((((.(((((((...))--)))))((((((((((.((((((((....))))).)))....)))))..))))).(((((((....)).)))))))))))))))........ ( -48.00) >consensus GCGCGAAGCUCGGUUUGGAGAGCGUCU__CUCCAGCUUUGUCGAGCUUUGUGCCCAUGCGUGCGAGAUCUUCGCCGGAAAGCUCUGACCAAUUCGGAAUCAGGCUGAGAUUUCAAUCGCG ((((((((((((.(((((((.........))))))....).))))))))))))....(((...((((((((.(((.((....(((((.....))))).)).))).))))))))...))). (-37.70 = -39.70 + 2.00)

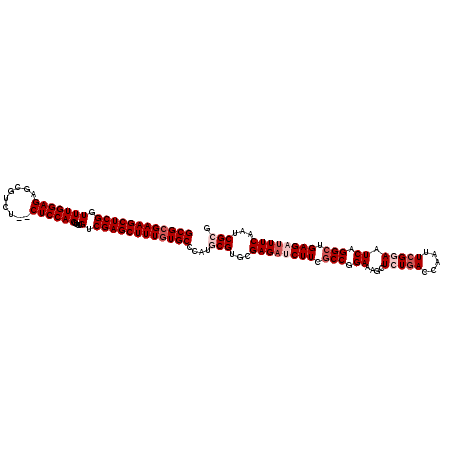

| Location | 8,032,692 – 8,032,810 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -31.96 |
| Energy contribution | -32.24 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8032692 118 - 23771897 CGCGAUUGAAAUCUCAGCCUGAUUCCGAAUUUGUCAGAGCUUUCCGGCGAAGAUCUCGCACGCAUGGGCACAAAGCUCGACAAAGCUGGAG--AGACGCUCUCCAAACCGAGCUUCGCGC (((((......((((........((((..((((((.(((((((...((((.....))))..((....))..)))))))))))))..)))))--))).((((........)))).))))). ( -39.80) >DroSec_CAF1 4142 118 - 1 CGCGAUUGAAAUCUCAGCCUGAUUUCGAAUUGGUCAGAGCUUUCCGGCGAAGAUCUCGCACGCAUGGGCACAAAGCUCGACAAAGCUGGAG--CGACGCUCUCCAAACCGAGCUUCGCGC .((..((((((((.......))))))))....(((.(((((((...((((.....))))..((....))..))))))))))...))....(--(((.((((........)))).)))).. ( -40.00) >DroSim_CAF1 4037 118 - 1 CGCGAUUGAAAUCUCAGCCUGAUUUCGAAUUGGUCAGAGCUUUCCGGCGAAGAUCUCGCACGCAUGGGCACAAAGCUCGACAAAGCUGGAG--AGACGCUCUCCAAACCGAGCUUCGCGC (((((((((((((.......)))))))).(((((...((((((.((((((.....)))).))..(((((.....)))))..))))))((((--((...))))))..)))))...))))). ( -42.10) >DroEre_CAF1 4093 120 - 1 CUCGAUUGAACGCUCAGCCUGAUUCCGAAUGGGUCAGAGCUUUCCGGUGAAGAUCUCGUACGCAUGGGCACAAAGCUCGCCAAAGCUGGAGAGAGACGCUCUCCAAACCGAGCUUCGCGC ..........((...((((((((((.....))))))).)))...))((((((..((((...((...(((.........)))...))(((((((.....)))))))...)))))))))).. ( -39.40) >DroYak_CAF1 4154 118 - 1 CUCGAUUGAAAGCUCAGCCUGACUCCCAAUGGGUCAGAGCUUUCCGGCGAAGAUCUCAUACGCAUGGGCAUAAAGCUCGCCAAAGCUGGAG--AGACGCUCUCCAAACCGAGCUUCGCGC .......((((((((....((((((.....))))))))))))))..((((((..(((....((...(((.........)))...))(((((--((...)))))))....))))))))).. ( -43.30) >consensus CGCGAUUGAAAUCUCAGCCUGAUUCCGAAUUGGUCAGAGCUUUCCGGCGAAGAUCUCGCACGCAUGGGCACAAAGCUCGACAAAGCUGGAG__AGACGCUCUCCAAACCGAGCUUCGCGC (((((........(((((((((((.......)))))).))).((((((((.....)).......(((((.....))))).....))))))....)).((((........)))).))))). (-31.96 = -32.24 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:16 2006