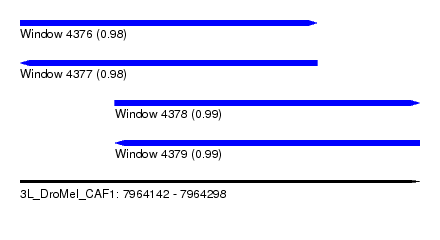
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,964,142 – 7,964,298 |
| Length | 156 |
| Max. P | 0.991547 |

| Location | 7,964,142 – 7,964,258 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -22.30 |
| Energy contribution | -22.97 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7964142 116 + 23771897 UGCAACUGCAACAGCAACAGCAACAGCAACUUUGGCAACUGCAACUCCGCAGCAACGGCAGUA-AACUUACUGCCAUUGUCUGGAUUCCCUCGGCUGCAUCCUCCACAGCGUUUGCC .((((.(((....((....))..((((......(((((((((......))))....(((((((-....))))))).))))).((....))...))))...........))).)))). ( -34.40) >DroSec_CAF1 7324 105 + 1 UGCAACUGCAGCA------------GCAACUUUUGCAACUGCAACUCCGCAGCAACGGCAGAAAAACUUACUGCCAUUGUCUGGAUUCCCUCGGCUGCAUCCUCCACAGCGUUUGCC .((((.(((((..------------(((.....)))..))))).....(((((...(((((.........))))).......((....))...)))))..............)))). ( -30.10) >DroEre_CAF1 7403 100 + 1 ----------------ACUGCAAGAGCAACUUUUGCAACUGCAACUCCGCAGCAACGGCAGUA-AACUUACUGCCAUUGUCCGGAUUCCCUCGACUGCAUCCUCCUCAGCGUUUGCC ----------------..((((((((...))))))))...((((...(((.((((.(((((((-....))))))).))))..((((.(........).))))......))).)))). ( -29.90) >consensus UGCAACUGCA_CA___AC_GCAA_AGCAACUUUUGCAACUGCAACUCCGCAGCAACGGCAGUA_AACUUACUGCCAUUGUCUGGAUUCCCUCGGCUGCAUCCUCCACAGCGUUUGCC .........................((((...((((....))))...(((.((((.(((((((.....))))))).))))..((((.(........).))))......))).)))). (-22.30 = -22.97 + 0.67)

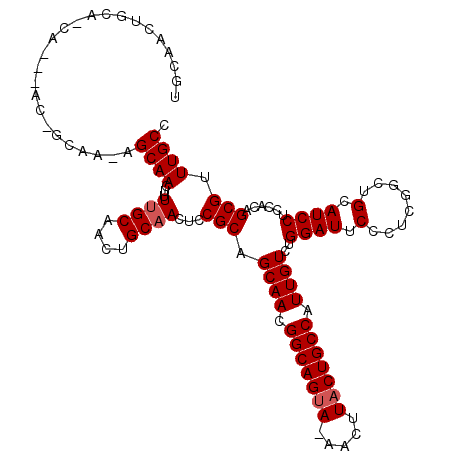
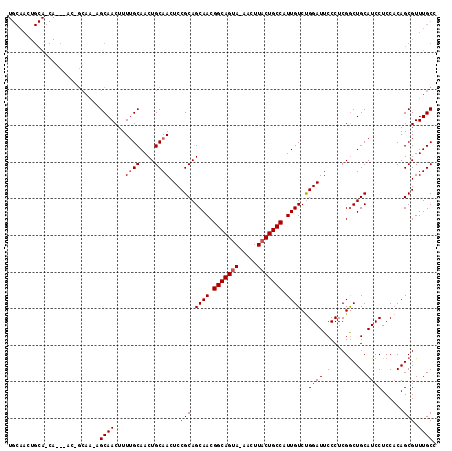
| Location | 7,964,142 – 7,964,258 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -41.27 |
| Consensus MFE | -30.79 |
| Energy contribution | -30.90 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7964142 116 - 23771897 GGCAAACGCUGUGGAGGAUGCAGCCGAGGGAAUCCAGACAAUGGCAGUAAGUU-UACUGCCGUUGCUGCGGAGUUGCAGUUGCCAAAGUUGCUGUUGCUGUUGCUGUUGCAGUUGCA ((((...((((..(....((((((...((....))....((((((((((....-))))))))))))))))...)..))))))))...((.(((((.((....))....))))).)). ( -46.80) >DroSec_CAF1 7324 105 - 1 GGCAAACGCUGUGGAGGAUGCAGCCGAGGGAAUCCAGACAAUGGCAGUAAGUUUUUCUGCCGUUGCUGCGGAGUUGCAGUUGCAAAAGUUGC------------UGCUGCAGUUGCA .((((((.(((..(.((((....((...)).))))...(((((((((.........))))))))))..))).))((((((.(((.....)))------------.)))))).)))). ( -39.80) >DroEre_CAF1 7403 100 - 1 GGCAAACGCUGAGGAGGAUGCAGUCGAGGGAAUCCGGACAAUGGCAGUAAGUU-UACUGCCGUUGCUGCGGAGUUGCAGUUGCAAAAGUUGCUCUUGCAGU---------------- .......(((((((((.((((((.(..(.((.((((.((((((((((((....-))))))))))).).)))).)).).)))))).....).))))).))))---------------- ( -37.20) >consensus GGCAAACGCUGUGGAGGAUGCAGCCGAGGGAAUCCAGACAAUGGCAGUAAGUU_UACUGCCGUUGCUGCGGAGUUGCAGUUGCAAAAGUUGCU_UUGC_GU___UG_UGCAGUUGCA .(((...((((..(....((((((...((....))....((((((((((.....))))))))))))))))...)..))))))).................................. (-30.79 = -30.90 + 0.11)

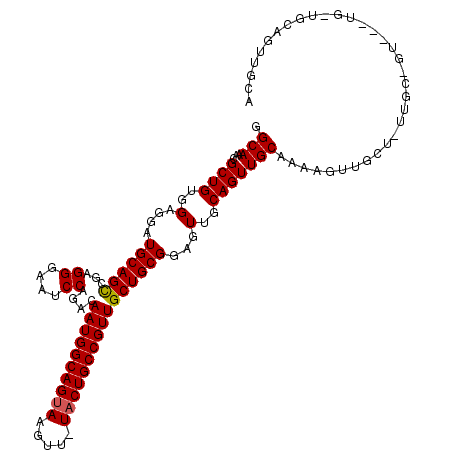
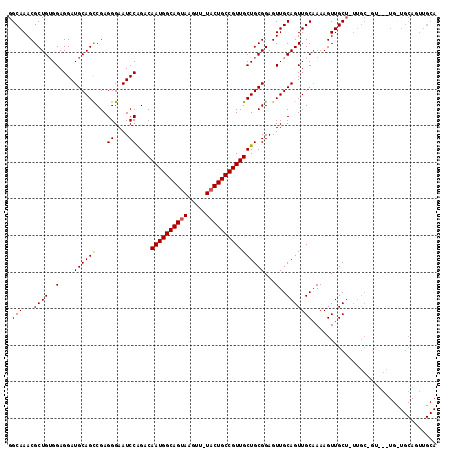
| Location | 7,964,179 – 7,964,298 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -38.69 |
| Energy contribution | -39.13 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7964179 119 + 23771897 ACUGCAACUCCGCAGCAACGGCAGUA-AACUUACUGCCAUUGUCUGGAUUCCCUCGGCUGCAUCCUCCACAGCGUUUGCCGCUGUUUAUGCUAAAUGAGGGUUAACGCAAAACUUAAAGA ..(((...((((..((((.(((((((-....))))))).)))).))))..((((((...((((.....((((((.....))))))..))))....)))))).....)))........... ( -41.10) >DroSec_CAF1 7349 120 + 1 ACUGCAACUCCGCAGCAACGGCAGAAAAACUUACUGCCAUUGUCUGGAUUCCCUCGGCUGCAUCCUCCACAGCGUUUGCCGCUGUUUAUGCUAAAUGAGGGUUAACGCAAAACUUAAAGA ..(((...((((..((((.(((((.........))))).)))).))))..((((((...((((.....((((((.....))))))..))))....)))))).....)))........... ( -37.60) >DroEre_CAF1 7424 119 + 1 ACUGCAACUCCGCAGCAACGGCAGUA-AACUUACUGCCAUUGUCCGGAUUCCCUCGACUGCAUCCUCCUCAGCGUUUGCCGCUGUUUAUGCUAAAUGAGGGUUAACGCAAAACUUGAAGA ..(((...((((..((((.(((((((-....))))))).)))).))))..((((((...((((......(((((.....)))))...))))....)))))).....)))........... ( -40.70) >consensus ACUGCAACUCCGCAGCAACGGCAGUA_AACUUACUGCCAUUGUCUGGAUUCCCUCGGCUGCAUCCUCCACAGCGUUUGCCGCUGUUUAUGCUAAAUGAGGGUUAACGCAAAACUUAAAGA ..(((...((((..((((.(((((((.....))))))).)))).))))..((((((...((((.....((((((.....))))))..))))....)))))).....)))........... (-38.69 = -39.13 + 0.45)

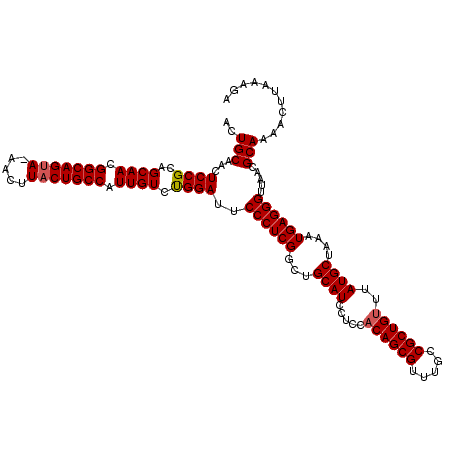
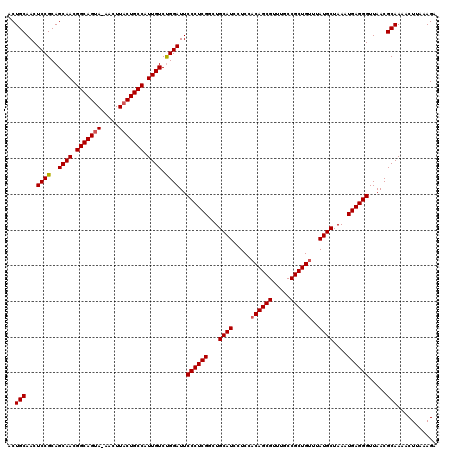
| Location | 7,964,179 – 7,964,298 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -45.40 |
| Consensus MFE | -43.45 |
| Energy contribution | -43.90 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7964179 119 - 23771897 UCUUUAAGUUUUGCGUUAACCCUCAUUUAGCAUAAACAGCGGCAAACGCUGUGGAGGAUGCAGCCGAGGGAAUCCAGACAAUGGCAGUAAGUU-UACUGCCGUUGCUGCGGAGUUGCAGU ..........(((((....(((((.....((((..((((((.....)))))).....))))....)))))..(((((.(((((((((((....-)))))))))))))).))...))))). ( -46.30) >DroSec_CAF1 7349 120 - 1 UCUUUAAGUUUUGCGUUAACCCUCAUUUAGCAUAAACAGCGGCAAACGCUGUGGAGGAUGCAGCCGAGGGAAUCCAGACAAUGGCAGUAAGUUUUUCUGCCGUUGCUGCGGAGUUGCAGU ..........(((((....(((((.....((((..((((((.....)))))).....))))....)))))..(((((.(((((((((.........)))))))))))).))...))))). ( -43.20) >DroEre_CAF1 7424 119 - 1 UCUUCAAGUUUUGCGUUAACCCUCAUUUAGCAUAAACAGCGGCAAACGCUGAGGAGGAUGCAGUCGAGGGAAUCCGGACAAUGGCAGUAAGUU-UACUGCCGUUGCUGCGGAGUUGCAGU ..........(((((....(((((..((.((((...(((((.....)))))......))))))..)))))..((((.((((((((((((....-))))))))))).).))))..))))). ( -46.70) >consensus UCUUUAAGUUUUGCGUUAACCCUCAUUUAGCAUAAACAGCGGCAAACGCUGUGGAGGAUGCAGCCGAGGGAAUCCAGACAAUGGCAGUAAGUU_UACUGCCGUUGCUGCGGAGUUGCAGU ..........(((((....(((((.....((((..((((((.....)))))).....))))....)))))..(((((.(((((((((((.....)))))))))))))).))...))))). (-43.45 = -43.90 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:06 2006