
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,962,711 – 7,962,871 |
| Length | 160 |
| Max. P | 0.999948 |

| Location | 7,962,711 – 7,962,831 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -35.94 |
| Consensus MFE | -32.32 |
| Energy contribution | -32.92 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7962711 120 - 23771897 UUUUCGUUUUGGUGGCACUUGGUUGACUAUUAUUUUUCAUUUGGGGCGCUCGCAGCUCGCAGGCGGUCCAAGUGAAAAUUAUUUCAAGAUACUGGUAAAACGCAAAGGCACACGCAUAGA (((((((((((.(((..(((((........((.((((((((((((.(((..((.....))..))).)))))))))))).))..)))))...))).))))))).))))((....))..... ( -35.90) >DroSec_CAF1 5832 120 - 1 UUUUCGUUUUGGUGGCACUUAGUUGACUAUUAUUUUUCAUUUGGGGCGCUCGCAGCUCGCAGGCGGUCCAAGUGAAAAUUAUUUCAAGAUACUGGUAAAACGCAAAGGCACACGCAUAGA (((((((((((.(((.......((((....((.((((((((((((.(((..((.....))..))).)))))))))))).))..))))....))).))))))).))))((....))..... ( -34.00) >DroSim_CAF1 5845 120 - 1 UUUUCGUUUUGGUGGCACUUGGUUGACUAUUAUUUUUCAUUUGGGGCGCUCGCAGCUCGCAGGCGGUCCAAGUGAAAAUUAUUUCAAGAUACUGGUAAAACGCAAAGGCACACGCAUAGA (((((((((((.(((..(((((........((.((((((((((((.(((..((.....))..))).)))))))))))).))..)))))...))).))))))).))))((....))..... ( -35.90) >DroEre_CAF1 5995 119 - 1 -UUACGUUUUGGUGGCACUUGGUUGACUAUUAUUUUUCAUUUGGGGCGCUCGCAGCUCGCAGGCGGUCCAAGUGAAAAUUAUUUCAAGAUACUGGUAAAACGCACAGGCACACGCACAGC -....(((.(((((((.(((((........((.((((((((((((.(((..((.....))..))).)))))))))))).))..)))))...(((((.....)).))))).))).)).))) ( -37.30) >DroYak_CAF1 6369 112 - 1 -UUACGUUUUGGUGGCACUUGGUUGACUAUUAUUUUUCAUUUGGGCCGCU-------CGCAGGCGGUCCAAGUGAAAAUUAUUUCAAGAUACUGGUAAAACGCAAAGGCACACGCAUAGA -...(((((((.(((..(((((........((.(((((((((((((((((-------....))))))))))))))))).))..)))))...))).))))))).....((....))..... ( -36.60) >consensus UUUUCGUUUUGGUGGCACUUGGUUGACUAUUAUUUUUCAUUUGGGGCGCUCGCAGCUCGCAGGCGGUCCAAGUGAAAAUUAUUUCAAGAUACUGGUAAAACGCAAAGGCACACGCAUAGA ....(((((((.(((..(((((........((.((((((((((((.((((.((.....)).)))).)))))))))))).))..)))))...))).))))))).....((....))..... (-32.32 = -32.92 + 0.60)

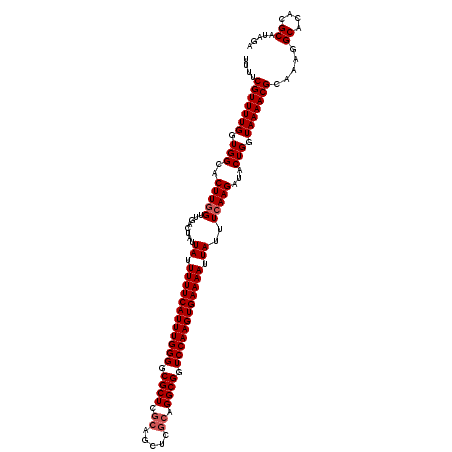
| Location | 7,962,751 – 7,962,871 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.66 |
| Mean single sequence MFE | -39.93 |
| Consensus MFE | -35.86 |
| Energy contribution | -36.50 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.77 |
| SVM RNA-class probability | 0.999948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7962751 120 - 23771897 CUCAGUUCACCAAUUCCAACAUAGAACCCAUGGAAGAUUUUUUUCGUUUUGGUGGCACUUGGUUGACUAUUAUUUUUCAUUUGGGGCGCUCGCAGCUCGCAGGCGGUCCAAGUGAAAAUU ...((((.(((((..(((...((((((....(((((....))))))))))).)))...))))).)))).....((((((((((((.(((..((.....))..))).)))))))))))).. ( -39.30) >DroSec_CAF1 5872 120 - 1 CUCAGUCCACCAAUCCCAACAUAGAACCCAUGGAAGCUUUUUUUCGUUUUGGUGGCACUUAGUUGACUAUUAUUUUUCAUUUGGGGCGCUCGCAGCUCGCAGGCGGUCCAAGUGAAAAUU ((.((((((((((..(...(((.......)))((((.....)))))..))))))).))).))...........((((((((((((.(((..((.....))..))).)))))))))))).. ( -38.60) >DroSim_CAF1 5885 120 - 1 CUCAGUCCACCAAUCCCAACAUAGAACCCAUGGAAGCUUUUUUUCGUUUUGGUGGCACUUGGUUGACUAUUAUUUUUCAUUUGGGGCGCUCGCAGCUCGCAGGCGGUCCAAGUGAAAAUU ...((((.(((((((........))..((((.(((((........))))).))))...))))).)))).....((((((((((((.(((..((.....))..))).)))))))))))).. ( -42.10) >DroEre_CAF1 6035 119 - 1 CCCAGUUCACCAAUCCCAACAUAGAAUCCAUGGAAGCUUU-UUACGUUUUGGUGGCACUUGGUUGACUAUUAUUUUUCAUUUGGGGCGCUCGCAGCUCGCAGGCGGUCCAAGUGAAAAUU ...((((.(((((((........))..((((.(((((...-....))))).))))...))))).)))).....((((((((((((.(((..((.....))..))).)))))))))))).. ( -39.90) >DroYak_CAF1 6409 112 - 1 CCCAGUUCACCAAUCCCAACAUACAACCCAUGGAAGCUUU-UUACGUUUUGGUGGCACUUGGUUGACUAUUAUUUUUCAUUUGGGCCGCU-------CGCAGGCGGUCCAAGUGAAAAUU ...((((.(((((..............((((.(((((...-....))))).))))...))))).)))).....(((((((((((((((((-------....))))))))))))))))).. ( -39.73) >consensus CUCAGUUCACCAAUCCCAACAUAGAACCCAUGGAAGCUUUUUUUCGUUUUGGUGGCACUUGGUUGACUAUUAUUUUUCAUUUGGGGCGCUCGCAGCUCGCAGGCGGUCCAAGUGAAAAUU ...((((.(((((((........))..((((.(((((........))))).))))...))))).)))).....((((((((((((.((((.((.....)).)))).)))))))))))).. (-35.86 = -36.50 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:01 2006