| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 109,335 – 109,429 |
| Length | 94 |
| Max. P | 0.992054 |

| Location | 109,335 – 109,429 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -19.69 |
| Consensus MFE | -4.72 |
| Energy contribution | -6.28 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.24 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 109335 94 + 23771897 AAUAGCGUCGCACCACGCAAUACCGUGCACCAAAAAAGCACAGGUGUAGCAAAAUGUUCCC-------CAAAAAUGGGAAAAUGCAAAAUGUUAUAAAGGC .(((((((.(((....((.(((((((((.........)))).))))).))......(((((-------.......)))))..)))...)))))))...... ( -25.50) >DroSec_CAF1 44970 81 + 1 GAUAGCGCUGCACAAUGCAAUACCGUGCACCGAAGAAGCACAGGGGUCGCAAAAUGUUCGC-------GC-------------ACAAAAAGUUAUAAAGGC ....((((.(((...(((.(..((((((.........)))).))..).)))...)))..))-------))-------------.................. ( -19.40) >DroSim_CAF1 50675 81 + 1 GAUAGCGCUGCACCAUGCAAUACCGUGCACCGAAGAAGCACAGGGGUCGCAAAAUGUUCGC-------GC-------------ACAAAAAGUUAUAAAGGC ....((((.(((...(((.(..((((((.........)))).))..).)))...)))..))-------))-------------.................. ( -19.40) >DroEre_CAF1 67012 87 + 1 GAUAGCAUCGCACCCUGCAAUACCGUGCACCAAAAAAU-GCAGGGAUAGCAAAAUGUUUGCAAAAUGUGC-------------ACAAAAAUUUAGAAAGGC ....((((....(((((((..................)-))))))...((((.....)))).....))))-------------.................. ( -18.07) >DroYak_CAF1 62446 81 + 1 GAUAACAUCGCACCCAGCAAUACCGUGCACCAAAAAAG-UGACGGAUUGCAAAAAGUUUCC------CGC-------------ACAAAAAGUUAGAAAGGC .............((.(((((.((((.(((.......)-)))))))))))......((((.------.((-------------.......))..)))))). ( -16.10) >consensus GAUAGCGUCGCACCAUGCAAUACCGUGCACCAAAAAAGCACAGGGGUAGCAAAAUGUUCGC_______GC_____________ACAAAAAGUUAUAAAGGC ....(((..(((...(((.((.((((((.........)))).)).)).)))...))).)))........................................ ( -4.72 = -6.28 + 1.56)

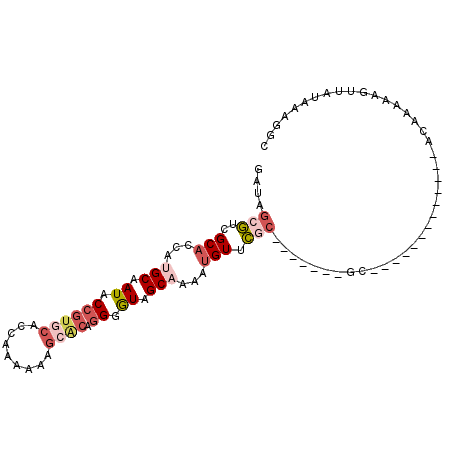
| Location | 109,335 – 109,429 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -10.18 |
| Energy contribution | -11.62 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 109335 94 - 23771897 GCCUUUAUAACAUUUUGCAUUUUCCCAUUUUUG-------GGGAACAUUUUGCUACACCUGUGCUUUUUUGGUGCACGGUAUUGCGUGGUGCGACGCUAUU ................((..(((((((....))-------)))))(((..(((...(((((..((.....))..)).)))...)))..)))....)).... ( -24.70) >DroSec_CAF1 44970 81 - 1 GCCUUUAUAACUUUUUGU-------------GC-------GCGAACAUUUUGCGACCCCUGUGCUUCUUCGGUGCACGGUAUUGCAUUGUGCAGCGCUAUC ..................-------------((-------((...(((..(((((..((.((((..(....).))))))..)))))..)))..)))).... ( -27.10) >DroSim_CAF1 50675 81 - 1 GCCUUUAUAACUUUUUGU-------------GC-------GCGAACAUUUUGCGACCCCUGUGCUUCUUCGGUGCACGGUAUUGCAUGGUGCAGCGCUAUC ..................-------------((-------((...(((..(((((..((.((((..(....).))))))..)))))..)))..)))).... ( -27.20) >DroEre_CAF1 67012 87 - 1 GCCUUUCUAAAUUUUUGU-------------GCACAUUUUGCAAACAUUUUGCUAUCCCUGC-AUUUUUUGGUGCACGGUAUUGCAGGGUGCGAUGCUAUC ................((-------------((((.....((((.....))))........(-(.....))))))))((((((((.....))))))))... ( -21.70) >DroYak_CAF1 62446 81 - 1 GCCUUUCUAACUUUUUGU-------------GCG------GGAAACUUUUUGCAAUCCGUCA-CUUUUUUGGUGCACGGUAUUGCUGGGUGCGAUGUUAUC .......((((...(((.-------------.((------(....))....(((((((((((-((.....)))).)))).)))))...)..))).)))).. ( -24.50) >consensus GCCUUUAUAACUUUUUGU_____________GC_______GCGAACAUUUUGCGACCCCUGUGCUUUUUUGGUGCACGGUAUUGCAUGGUGCGACGCUAUC ........................................(((..(((..(((((..((((((((.....)))))).))..)))))..)))...))).... (-10.18 = -11.62 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:15 2006