
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,958,173 – 7,958,316 |
| Length | 143 |
| Max. P | 0.994795 |

| Location | 7,958,173 – 7,958,276 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.51 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -24.92 |
| Energy contribution | -24.80 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7958173 103 + 23771897 ----UGCCUUUGUUUUGGCCAGUUACCCGAGUUCCCUUGU-------------UCCCUGGCAACAGGCUCAUUUGGCCAAGAACAAUUUAGAGCCUUUUAAUAGCCUUUAUGUUUUAUUG ----.....(((((((((((((.....((((....)))).-------------..((((....)))).....)))))).)))))))..(((((.((......)).))))).......... ( -26.30) >DroSec_CAF1 3667 103 + 1 ----UGCCUUUGUUUUGGCCAGUUACCCGAGUUCCCUUGU-------------UCCCUGGUAACAGGCUCAUUUGGCCAAGAACAAUUUAGAGCCUUUUAAUGGCCUUUAUGUUUUAUUG ----............(((((((((((((((....)))).-------------.....))))))((((((..(((........)))....)))))).....))))).............. ( -26.90) >DroSim_CAF1 3668 103 + 1 ----UGCCUUUGUUUUGGCCAGUUACCCGAGUUCCCUUGU-------------UCCCUGGUAACAGGCUCAUUUGGCUAAGAACAAUUUAGAGCCUUUUAAUGGCCUUUAUGUUUUAUUG ----............(((((((((((((((....)))).-------------.....))))))((((((..(((........)))....)))))).....))))).............. ( -26.90) >DroEre_CAF1 3828 114 + 1 CCUUCGCCUUUGUUUUGGCCAGUUACCCGAGUUCCCUUGUCGCCCUGCC-----UCCUGGUAACAGGCUCAUUUGGCCAGGAACAGUUUAGAGCCU-UUAGUGGCCUUUAUGUUUUAUUG ...............(((((((.....((((....))))..(((.((((-----....))))...)))....)))))))((((((....((.(((.-.....)))))...)))))).... ( -30.20) >DroYak_CAF1 4057 120 + 1 CCUUUGCCUUUGUUUUGGCCAGUUACCCGAGUUCCCUUGCCACCUUGUUCCCUUCCCUGGUAACAGGCUCAUUUGGCCAAGAACAAUUUAGAGCCUUUUAAUGGCCUUUAUGUUUUAUUG .........(((((((((((((.....((((....))))....((((((.((......)).)))))).....)))))).)))))))..(((((((.......)).))))).......... ( -27.90) >consensus ____UGCCUUUGUUUUGGCCAGUUACCCGAGUUCCCUUGU_____________UCCCUGGUAACAGGCUCAUUUGGCCAAGAACAAUUUAGAGCCUUUUAAUGGCCUUUAUGUUUUAUUG ...............(((((((.....((((....))))................((((....)))).....)))))))((((((....((.(((.......)))))...)))))).... (-24.92 = -24.80 + -0.12)


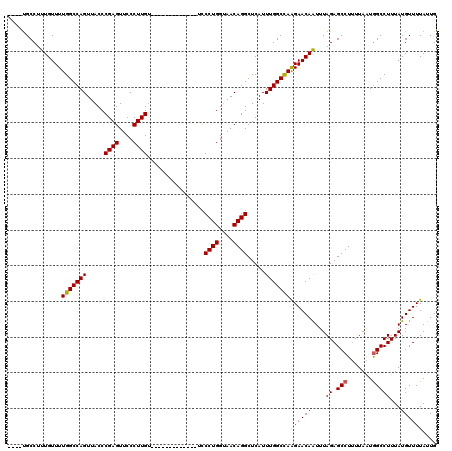
| Location | 7,958,173 – 7,958,276 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.51 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -25.44 |
| Energy contribution | -25.32 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7958173 103 - 23771897 CAAUAAAACAUAAAGGCUAUUAAAAGGCUCUAAAUUGUUCUUGGCCAAAUGAGCCUGUUGCCAGGGA-------------ACAAGGGAACUCGGGUAACUGGCCAAAACAAAGGCA---- ..............(((((.....((((((....(((........)))..))))))((((((.(((.-------------.........))).)))))))))))............---- ( -25.40) >DroSec_CAF1 3667 103 - 1 CAAUAAAACAUAAAGGCCAUUAAAAGGCUCUAAAUUGUUCUUGGCCAAAUGAGCCUGUUACCAGGGA-------------ACAAGGGAACUCGGGUAACUGGCCAAAACAAAGGCA---- ..............(((((.....((((((....(((........)))..))))))((((((.(((.-------------.........))).)))))))))))............---- ( -27.70) >DroSim_CAF1 3668 103 - 1 CAAUAAAACAUAAAGGCCAUUAAAAGGCUCUAAAUUGUUCUUAGCCAAAUGAGCCUGUUACCAGGGA-------------ACAAGGGAACUCGGGUAACUGGCCAAAACAAAGGCA---- ..............(((((.....((((((....(((........)))..))))))((((((.(((.-------------.........))).)))))))))))............---- ( -27.70) >DroEre_CAF1 3828 114 - 1 CAAUAAAACAUAAAGGCCACUAA-AGGCUCUAAACUGUUCCUGGCCAAAUGAGCCUGUUACCAGGA-----GGCAGGGCGACAAGGGAACUCGGGUAACUGGCCAAAACAAAGGCGAAGG ..............(((((....-..((((......((((((.(((...((..((((....)))).-----..)).))).....))))))..))))...)))))................ ( -30.00) >DroYak_CAF1 4057 120 - 1 CAAUAAAACAUAAAGGCCAUUAAAAGGCUCUAAAUUGUUCUUGGCCAAAUGAGCCUGUUACCAGGGAAGGGAACAAGGUGGCAAGGGAACUCGGGUAACUGGCCAAAACAAAGGCAAAGG .............(((((.......))).))...(((((.(((((((..(((((((((((((..(........)..)))))).)))...))))......))))))))))))......... ( -31.10) >consensus CAAUAAAACAUAAAGGCCAUUAAAAGGCUCUAAAUUGUUCUUGGCCAAAUGAGCCUGUUACCAGGGA_____________ACAAGGGAACUCGGGUAACUGGCCAAAACAAAGGCA____ ..............(((((.....((((((......((.....)).....))))))((((((.(((.......................))).)))))))))))................ (-25.44 = -25.32 + -0.12)


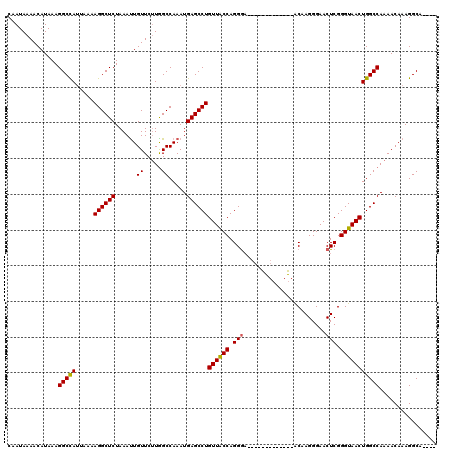
| Location | 7,958,209 – 7,958,316 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.60 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -24.12 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7958209 107 + 23771897 -------------UCCCUGGCAACAGGCUCAUUUGGCCAAGAACAAUUUAGAGCCUUUUAAUAGCCUUUAUGUUUUAUUGGUACACACACAUAAUGUGUACAUUGUGGUUACUUUGGAAC -------------..((((....)))).(((..((((((((((((...(((((.((......)).)))))))))))....(((((((.......)))))))....))))))...)))... ( -27.60) >DroSec_CAF1 3703 105 + 1 -------------UCCCUGGUAACAGGCUCAUUUGGCCAAGAACAAUUUAGAGCCUUUUAAUGGCCUUUAUGUUUUAUUGGUACAC--ACAUAAUGUGUACAUUGUGGUUACUUUGGAAC -------------(((..((((((.((((.....)))).((((((....((.(((.......)))))...))))))....((((((--(.....)))))))......))))))..))).. ( -32.20) >DroSim_CAF1 3704 105 + 1 -------------UCCCUGGUAACAGGCUCAUUUGGCUAAGAACAAUUUAGAGCCUUUUAAUGGCCUUUAUGUUUUAUUGGUACAC--ACAUAAUGUGUACAUUGUGGUUACUUUGGAAC -------------(((..((((((((((.((((.((((..((.....))..))))....)))))))).............((((((--(.....)))))))......))))))..))).. ( -31.70) >DroEre_CAF1 3868 109 + 1 CGCCCUGCC-----UCCUGGUAACAGGCUCAUUUGGCCAGGAACAGUUUAGAGCCU-UUAGUGGCCUUUAUGUUUUAUUGGUACA----CAUGAUGUGUGCAUUGUGGCUACAUUG-AAC .(((.((((-----....))))...)))(((..((((((((((((....((.(((.-.....)))))...))))))....(((((----(.....))))))....))))))...))-).. ( -35.70) >DroYak_CAF1 4097 115 + 1 CACCUUGUUCCCUUCCCUGGUAACAGGCUCAUUUGGCCAAGAACAAUUUAGAGCCUUUUAAUGGCCUUUAUGUUUUAUUGGUACA----CAUAAUGUGUACAUUGUGGUUACUUUG-AAC ...((((((.((......)).)))))).(((..((((((((((((....((.(((.......)))))...))))))....(((((----(.....))))))....))))))...))-).. ( -31.10) >consensus _____________UCCCUGGUAACAGGCUCAUUUGGCCAAGAACAAUUUAGAGCCUUUUAAUGGCCUUUAUGUUUUAUUGGUACAC__ACAUAAUGUGUACAUUGUGGUUACUUUGGAAC .............(((..((((((((((((..(((........)))....)))))).......(((..((.....))..))).......(((((((....)))))))))))))..))).. (-24.12 = -25.00 + 0.88)

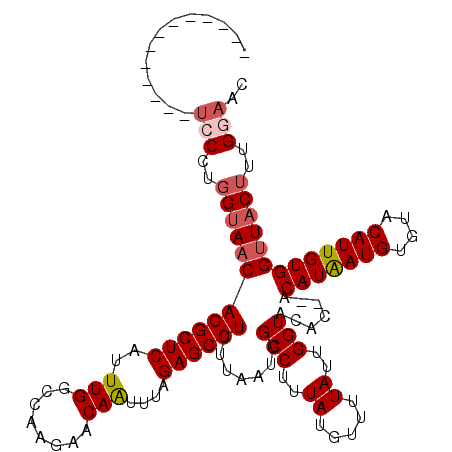

| Location | 7,958,209 – 7,958,316 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.60 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7958209 107 - 23771897 GUUCCAAAGUAACCACAAUGUACACAUUAUGUGUGUGUACCAAUAAAACAUAAAGGCUAUUAAAAGGCUCUAAAUUGUUCUUGGCCAAAUGAGCCUGUUGCCAGGGA------------- ..(((...(((((......((((((((.....)))))))).............(((((.......((((.............)))).....))))))))))...)))------------- ( -26.92) >DroSec_CAF1 3703 105 - 1 GUUCCAAAGUAACCACAAUGUACACAUUAUGU--GUGUACCAAUAAAACAUAAAGGCCAUUAAAAGGCUCUAAAUUGUUCUUGGCCAAAUGAGCCUGUUACCAGGGA------------- ..(((...(((((......((((((((...))--)))))).............((((((((....((((.............)))).)))).)))))))))...)))------------- ( -26.32) >DroSim_CAF1 3704 105 - 1 GUUCCAAAGUAACCACAAUGUACACAUUAUGU--GUGUACCAAUAAAACAUAAAGGCCAUUAAAAGGCUCUAAAUUGUUCUUAGCCAAAUGAGCCUGUUACCAGGGA------------- ..(((...(((((......((((((((...))--)))))).............((((((((....((((.............)))).)))).)))))))))...)))------------- ( -26.32) >DroEre_CAF1 3868 109 - 1 GUU-CAAUGUAGCCACAAUGCACACAUCAUG----UGUACCAAUAAAACAUAAAGGCCACUAA-AGGCUCUAAACUGUUCCUGGCCAAAUGAGCCUGUUACCAGGA-----GGCAGGGCG ...-.......(((....((((((.....))----))))..............(((((.....-.))).))...((((((((((.((........))...))))))-----.))))))). ( -27.70) >DroYak_CAF1 4097 115 - 1 GUU-CAAAGUAACCACAAUGUACACAUUAUG----UGUACCAAUAAAACAUAAAGGCCAUUAAAAGGCUCUAAAUUGUUCUUGGCCAAAUGAGCCUGUUACCAGGGAAGGGAACAAGGUG ...-.......(((.....((((((.....)----))))).............(((((.......))).))...((((((((..((...((..........)).))..))))))))))). ( -25.50) >consensus GUUCCAAAGUAACCACAAUGUACACAUUAUGU__GUGUACCAAUAAAACAUAAAGGCCAUUAAAAGGCUCUAAAUUGUUCUUGGCCAAAUGAGCCUGUUACCAGGGA_____________ ..(((...(((((......((((((.........)))))).............((((((((....((((.............)))).)))).)))))))))...)))............. (-17.98 = -18.74 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:52 2006