
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,906,001 – 7,906,111 |
| Length | 110 |
| Max. P | 0.914978 |

| Location | 7,906,001 – 7,906,111 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.63 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -21.75 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
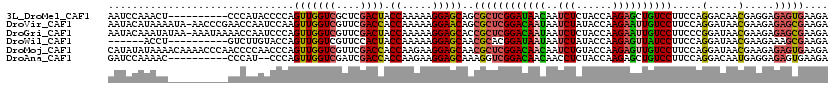
>3L_DroMel_CAF1 7906001 110 + 23771897 UCUUCACUCUCCUCGUUGUCCUGGAAGGACAGCUCUUGGUAGAGAUUGUUAUCCGAGCGCUGCUCCUUUUUGGUAGUCGAGCGACCAACUGGGGUAUGGG----------AGUUUGGAUU (((..((((((((((....((..((((((((((.((((((((......))).))))).)))).))))))..))....))))..(((......)))..)))----------)))..))).. ( -40.40) >DroVir_CAF1 7671 119 + 1 UCUUCGCUCUCUUCGUUAUCCUGGAAGGACAAUUCUUGGUAUAGAUUAUUGUCCGAGCGCUGUUCCUUUUUGGUGGUCGAACGACCAACUUGGAUUGGUUCGGGUU-UAUUUUAUGUAUU ....(((((..((((......)))).((((((((((......)))..))))))))))))(((..((..((..((((((....)))))..)..))..))..)))...-............. ( -32.70) >DroGri_CAF1 7333 119 + 1 UCUUCGCUCUCUUCGUUAUCCGGGAAGGACAAUUCUUGGUAGAGAUUAUUGUCCGAGCGGUGCUCCUUUUUGGUAGUCGAACGACCAACUGGGAUUGGUUUUAUUU-UUAUAUUUGUAUU ...(((((((((.((.....))))).(((((((((((....))))..)))))))))))))...((((..(((((.(.....).)))))..))))............-............. ( -30.60) >DroWil_CAF1 6320 104 + 1 UCUUCGCUUUCUUCGUUAUCCUGGAAGGAUAACUCUUGGUAUAGAUUAUUAUCCGUGCGUUGCUCCUUUUUGGUAGUGGAACGACCAACUGGUACAAGAC----------AGGU------ .....((((.....(((((((.....)))))))((((((((..(((....)))..))).(..((((.....)).))..)....(((....))).))))).----------))))------ ( -25.00) >DroMoj_CAF1 7370 120 + 1 UCUUCACUCUCUUCGUUAUCCUGGAAGGACAACUCUUGGUACAGAUUGUUGUCCGAGCGUUGCUCCUUCUUGGUGGUCGAACGACCAACUGGGUUGGGGUUGGGUUUUGUUUUAUAUAUG .((..((((..((((....((.(((((((((((.(((((.((((....))))))))).)))).))))))).))....))))(((((.....)))))))))..))................ ( -40.20) >DroAna_CAF1 5125 108 + 1 UCUUCACUCUCCUCAUUGUCCUGGAAGGACAGCUCUUGGUAGAGGUUGUUGUCCGACCUUUGCUCCUUCUUGGUGGUCGAUCGACCAACUGGG--AUGGG----------GUUUUGGAUC ..((((....(((((((((((.....)))))).....(((((((((((.....)))))))))))...(((..((((((....)))).))..))--)))))----------)...)))).. ( -39.90) >consensus UCUUCACUCUCUUCGUUAUCCUGGAAGGACAACUCUUGGUAGAGAUUAUUGUCCGAGCGUUGCUCCUUUUUGGUAGUCGAACGACCAACUGGGAUAGGGU__________UUUUUGUAUU .........((((((....((..((((((((((.(((((.............))))).)))).))))))..))....)))).))((....))............................ (-21.75 = -22.30 + 0.56)



| Location | 7,906,001 – 7,906,111 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.63 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -18.02 |
| Energy contribution | -17.75 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7906001 110 - 23771897 AAUCCAAACU----------CCCAUACCCCAGUUGGUCGCUCGACUACCAAAAAGGAGCAGCGCUCGGAUAACAAUCUCUACCAAGAGCUGUCCUUCCAGGACAACGAGGAGAGUGAAGA ..((...(((----------(((..........(((((....)))))((...(((((.((((.((.((.((........)))).)).)))))))))...)).......)).))))...)) ( -27.60) >DroVir_CAF1 7671 119 - 1 AAUACAUAAAAUA-AACCCGAACCAAUCCAAGUUGGUCGUUCGACCACCAAAAAGGAACAGCGCUCGGACAAUAAUCUAUACCAAGAAUUGUCCUUCCAGGAUAACGAAGAGAGCGAAGA .............-............(((...(((((.(.....).)))))...)))....((((((((((((..(((......)))))))))).....(.....).....))))).... ( -25.30) >DroGri_CAF1 7333 119 - 1 AAUACAAAUAUAA-AAAUAAAACCAAUCCCAGUUGGUCGUUCGACUACCAAAAAGGAGCACCGCUCGGACAAUAAUCUCUACCAAGAAUUGUCCUUCCCGGAUAACGAAGAGAGCGAAGA .............-............(((...(((((.(.....).)))))...)))....((((((((((((..(((......))))))))))....((.....))....))))).... ( -27.00) >DroWil_CAF1 6320 104 - 1 ------ACCU----------GUCUUGUACCAGUUGGUCGUUCCACUACCAAAAAGGAGCAACGCACGGAUAAUAAUCUAUACCAAGAGUUAUCCUUCCAGGAUAACGAAGAAAGCGAAGA ------.(((----------((.((((.((..(((((.(.....).)))))...)).)))).))).)).......(((......)))(((((((.....))))))).............. ( -25.00) >DroMoj_CAF1 7370 120 - 1 CAUAUAUAAAACAAAACCCAACCCCAACCCAGUUGGUCGUUCGACCACCAAGAAGGAGCAACGCUCGGACAACAAUCUGUACCAAGAGUUGUCCUUCCAGGAUAACGAAGAGAGUGAAGA (((..............(((((.........)))))((.((((....((..((((((.((((.((.(((((......))).)).)).))))))))))..))....))))..))))).... ( -28.40) >DroAna_CAF1 5125 108 - 1 GAUCCAAAAC----------CCCAU--CCCAGUUGGUCGAUCGACCACCAAGAAGGAGCAAAGGUCGGACAACAACCUCUACCAAGAGCUGUCCUUCCAGGACAAUGAGGAGAGUGAAGA ..........----------....(--((((((((..(((((...(.((.....)).)....)))))..))))...((((....)))).(((((.....))))).)).)))......... ( -25.70) >consensus AAUACAAAAA__________ACCCUAUCCCAGUUGGUCGUUCGACCACCAAAAAGGAGCAACGCUCGGACAACAAUCUCUACCAAGAGUUGUCCUUCCAGGAUAACGAAGAGAGCGAAGA ...............................(((((((....)))).((.....)))))..((((((((((((..(((......)))))))))).....(.....).....))))).... (-18.02 = -17.75 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:38 2006