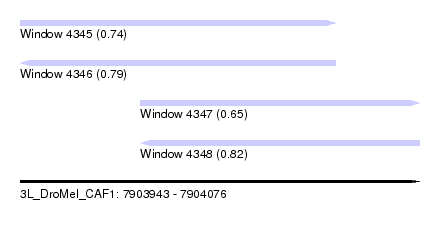
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,903,943 – 7,904,076 |
| Length | 133 |
| Max. P | 0.822955 |

| Location | 7,903,943 – 7,904,048 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.20 |
| Mean single sequence MFE | -46.82 |
| Consensus MFE | -38.05 |
| Energy contribution | -38.38 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7903943 105 + 23771897 UGUGGGCAGGGCUUCCUCCGCCGGUGUGACGAAACAUCGGCCCCCCUCCGAGACCGUCCGCAACGCGUUCCGUUGGGCGGUUGGAGGGGCUG-----UCCAU--AUA--G---AAAG (((((((((..........((((((((......)))))))).(((((((..(((((.((.(((((.....))))))))))))))))))))))-----)))))--)..--.---.... ( -55.80) >DroVir_CAF1 5661 103 + 1 UGUGGGCAGGGCUUCCUCCGCCUGUGUGACGAAACAUCGGCCCCCCUCCAAGCCCGUCGGCGCCGCGCUCCGACGAACGAUUGGAGUGGCUG-----UUCGGU--AA--G-----UA .(..(((.(((....))).)))..)....((((....(((((...((((((...((((((.((...)).)))))).....)))))).)))))-----))))..--..--.-----.. ( -37.70) >DroGri_CAF1 5269 108 + 1 UGUGGGCAGGGCUUCCUCCGCCUGUGUGACGAAACAUAGGCCCCCCUCCAAGGCCGUCAGCGACGCGUUCCGUCGGACGAUUGGAGGGGCUGCAAAUUUUUUA--UA--G-----CA .....((((..........((((((((......)))))))).(((((((((...((((..(((((.....))))))))).)))))))))))))..........--..--.-----.. ( -46.60) >DroMoj_CAF1 5440 103 + 1 UGUGGGCAGGACUUCCUCCGCCUGUGUGACGAAACAUCGGCCCCCCUCCAAGACCGUCAGCGACGCGUUCCGUCGGACGGUUGGAGGGGCUG-----UUGUGA--GA--G-----UA .(..(((.(((.....))))))..)...((...(((.(((((((..((((..((((((..(((((.....))))))))))))))))))))))-----.)))..--..--)-----). ( -45.70) >DroAna_CAF1 3080 102 + 1 UGUGGGCAGGGCUUCCUCCGCCGGUGUGACGAAACAUCGGCCCCCCUCCGAGACCGUCCGCGACGCGUUCCGUCGGGCGGUUGGAGGGGCUG-----UUUGGA----------AGAA ...........(((((...((((((((......)))))))).(((((((..(((((((((((........)).))))))))))))))))...-----...)))----------)).. ( -51.90) >DroPer_CAF1 3547 112 + 1 UGUGGGCAGGACUUCCUCCCCCGGUAUUACGAAACAUCGGCCCCCCUCCCAGCCCGUCGGCGAUGCGUUCCGUCGGGCGAUUGGAGGGGCUG-----UCACGAAACAGAGUAGAGAC ...(((.(((....))))))(((((.((....)).)))))..(((((((..(((((.(((.........))).)))))....)))))))(((-----(......))))......... ( -43.20) >consensus UGUGGGCAGGGCUUCCUCCGCCGGUGUGACGAAACAUCGGCCCCCCUCCAAGACCGUCAGCGACGCGUUCCGUCGGACGAUUGGAGGGGCUG_____UUCGGA__UA__G_____AA ....((.(((....)))))((((((((......)))))))).(((((((((...((((..(((((.....))))))))).)))))))))............................ (-38.05 = -38.38 + 0.34)


| Location | 7,903,943 – 7,904,048 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.20 |
| Mean single sequence MFE | -45.35 |
| Consensus MFE | -32.19 |
| Energy contribution | -31.92 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7903943 105 - 23771897 CUUU---C--UAU--AUGGA-----CAGCCCCUCCAACCGCCCAACGGAACGCGUUGCGGACGGUCUCGGAGGGGGGCCGAUGUUUCGUCACACCGGCGGAGGAAGCCCUGCCCACA ....---.--...--.(((.-----((((((((((.(((((((((((.....))))).)).))))...)))))))(((...(.(((((((.....))))))).).)))))).))).. ( -45.30) >DroVir_CAF1 5661 103 - 1 UA-----C--UU--ACCGAA-----CAGCCACUCCAAUCGUUCGUCGGAGCGCGGCGCCGACGGGCUUGGAGGGGGGCCGAUGUUUCGUCACACAGGCGGAGGAAGCCCUGCCCACA ..-----.--..--......-----...((.((((((..(((((((((.((...)).)))))))))))))))))((((.(..(((((.((.(......))).))))).).))))... ( -41.30) >DroGri_CAF1 5269 108 - 1 UG-----C--UA--UAAAAAAUUUGCAGCCCCUCCAAUCGUCCGACGGAACGCGUCGCUGACGGCCUUGGAGGGGGGCCUAUGUUUCGUCACACAGGCGGAGGAAGCCCUGCCCACA ..-----.--..--..........(((((((((((((((((((((((.....)))))..)))))..)))))))))(((...(.(((((((.....))))))).).)))))))..... ( -44.00) >DroMoj_CAF1 5440 103 - 1 UA-----C--UC--UCACAA-----CAGCCCCUCCAACCGUCCGACGGAACGCGUCGCUGACGGUCUUGGAGGGGGGCCGAUGUUUCGUCACACAGGCGGAGGAAGUCCUGCCCACA ..-----.--..--......-----....((((((((((((((((((.....)))))..))))))..)))))))((((.(((.(((((((.....)))))))...)))..))))... ( -45.20) >DroAna_CAF1 3080 102 - 1 UUCU----------UCCAAA-----CAGCCCCUCCAACCGCCCGACGGAACGCGUCGCGGACGGUCUCGGAGGGGGGCCGAUGUUUCGUCACACCGGCGGAGGAAGCCCUGCCCACA ((((----------(((...-----...(((((((.(((((((((((.....))))).)).))))...))))))).((((.(((......))).)))))))))))............ ( -49.40) >DroPer_CAF1 3547 112 - 1 GUCUCUACUCUGUUUCGUGA-----CAGCCCCUCCAAUCGCCCGACGGAACGCAUCGCCGACGGGCUGGGAGGGGGGCCGAUGUUUCGUAAUACCGGGGGAGGAAGUCCUGCCCACA (((......(((((....))-----)))(((((((....(((((.(((.........))).)))))..))))))))))((.((((....)))).))(((.(((....))).)))... ( -46.90) >consensus UU_____C__UA__UCAAAA_____CAGCCCCUCCAACCGCCCGACGGAACGCGUCGCCGACGGUCUUGGAGGGGGGCCGAUGUUUCGUCACACAGGCGGAGGAAGCCCUGCCCACA ............................(((((((..(((..(((((.....)))))....)))....))))))).((((.(((......))).))))(((((....))).)).... (-32.19 = -31.92 + -0.27)


| Location | 7,903,983 – 7,904,076 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -27.96 |
| Energy contribution | -28.02 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7903983 93 + 23771897 CCCCCCUCCGAGACCGUCCGCAACGCGUUCCGUUGGGCGGUUGGAGGGGCUGUCCAU-AUAG-AAAGGUUAACAAGUUAAUA-UAGGGCAUUCUG-G ..(((((((..(((((.((.(((((.....))))))))))))))))))).(((((((-((.(-(...(....)...)).)))-).))))).....-. ( -36.20) >DroSec_CAF1 3272 94 + 1 CCCCCCUCCGAGACCGUCCGCAACGCGUUCCGUUGGGCGGUUGGAGGGGCUGUCAGGGGGUG-AAAGGUUAACAAGUUAAUA-UAGGGCAUUCUG-G ..(((((((..(((((.((.(((((.....))))))))))))))))))).......((((((-....(((((....))))).-.....)))))).-. ( -36.00) >DroSim_CAF1 3284 94 + 1 CCCCCCUCCGAGACCGUCCGCAACGCGUUCCGUUGGGCGGUUGGAGGGGCUGUCAGGGGGUG-AAAGGUUAACAAGUUAAUA-UACGGCAUUCUG-G ..(((((((..(((((.((.(((((.....))))))))))))))))))).......((((((-....(((((....))))).-.....)))))).-. ( -36.00) >DroEre_CAF1 3529 93 + 1 CCCCCCUCCGAGACCGUCCGCAACGCGUUCCGUUGGGCGGCUGGAGGAGCUGUUCAU-AUAG-AGCGGUAAAUAUGUUAAUA-AAGGGCAUUCAG-A .((((((((.((.(((.((.(((((.....))))))))))))))))).(((((((..-...)-)))))).............-..))).......-. ( -34.90) >DroYak_CAF1 3302 95 + 1 CCCCCCUCCGAGACCGUCCGCAACGCGUUCCGUUGGGCGGUUGGAGGGGCUGUCCAU-AUAGAAGCGGUUAACAUGUUAAUACAAAAGUUUUCUA-A ..(((((((..(((((.((.(((((.....)))))))))))))))))))........-.(((((((.(((((....)))))......).))))))-. ( -33.20) >DroAna_CAF1 3120 91 + 1 CCCCCCUCCGAGACCGUCCGCGACGCGUUCCGUCGGGCGGUUGGAGGGGCUGUUUGGA-----AGAAGUUCAAAAGUUAAUU-GUCUGCAUGGUAAG (((((((((..(((((((((((........)).))))))))))))))))...((((((-----.....))))))........-........)).... ( -33.90) >consensus CCCCCCUCCGAGACCGUCCGCAACGCGUUCCGUUGGGCGGUUGGAGGGGCUGUCCAG_AUAG_AAAGGUUAACAAGUUAAUA_UAGGGCAUUCUG_G ..(((((((..(((((.((.(((((.....)))))))))))))))))))................................................ (-27.96 = -28.02 + 0.06)



| Location | 7,903,983 – 7,904,076 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -31.39 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.35 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7903983 93 - 23771897 C-CAGAAUGCCCUA-UAUUAACUUGUUAACCUUU-CUAU-AUGGACAGCCCCUCCAACCGCCCAACGGAACGCGUUGCGGACGGUCUCGGAGGGGGG (-(....((((.((-((((((....)))).....-.)))-).)).)).(((((((.(((((((((((.....))))).)).))))...))))))))) ( -31.70) >DroSec_CAF1 3272 94 - 1 C-CAGAAUGCCCUA-UAUUAACUUGUUAACCUUU-CACCCCCUGACAGCCCCUCCAACCGCCCAACGGAACGCGUUGCGGACGGUCUCGGAGGGGGG (-(...........-.......((((((......-.......))))))(((((((.(((((((((((.....))))).)).))))...))))))))) ( -30.92) >DroSim_CAF1 3284 94 - 1 C-CAGAAUGCCGUA-UAUUAACUUGUUAACCUUU-CACCCCCUGACAGCCCCUCCAACCGCCCAACGGAACGCGUUGCGGACGGUCUCGGAGGGGGG .-.......((...-.......((((((......-.......))))))(((((((.(((((((((((.....))))).)).))))...))))))))) ( -31.12) >DroEre_CAF1 3529 93 - 1 U-CUGAAUGCCCUU-UAUUAACAUAUUUACCGCU-CUAU-AUGAACAGCUCCUCCAGCCGCCCAACGGAACGCGUUGCGGACGGUCUCGGAGGGGGG .-.......(((..-......(((((........-..))-))).......(((((((((((((((((.....))))).)).))).)).)))))))). ( -30.20) >DroYak_CAF1 3302 95 - 1 U-UAGAAAACUUUUGUAUUAACAUGUUAACCGCUUCUAU-AUGGACAGCCCCUCCAACCGCCCAACGGAACGCGUUGCGGACGGUCUCGGAGGGGGG .-.....(((...(((....))).)))..((((((((..-..))).)))((((((.(((((((((((.....))))).)).))))...)))))))). ( -32.40) >DroAna_CAF1 3120 91 - 1 CUUACCAUGCAGAC-AAUUAACUUUUGAACUUCU-----UCCAAACAGCCCCUCCAACCGCCCGACGGAACGCGUCGCGGACGGUCUCGGAGGGGGG ....((.......(-((.......))).......-----.........(((((((.(((((((((((.....))))).)).))))...))))))))) ( -32.00) >consensus C_CAGAAUGCCCUA_UAUUAACUUGUUAACCUCU_CUAU_ACGGACAGCCCCUCCAACCGCCCAACGGAACGCGUUGCGGACGGUCUCGGAGGGGGG ................................................(((((((.(((((((((((.....))))).)).))))...))))))).. (-29.90 = -29.35 + -0.55)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:36 2006