| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,872,644 – 7,872,750 |
| Length | 106 |
| Max. P | 0.515833 |

| Location | 7,872,644 – 7,872,750 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.16 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -18.37 |
| Energy contribution | -17.93 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
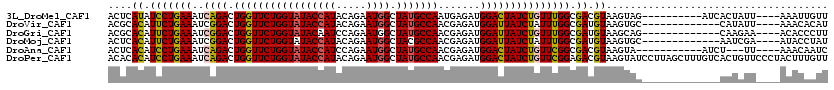
>3L_DroMel_CAF1 7872644 106 - 23771897 ACUCAUAUCCUGAAAUCAGACUGGUUCUGGUAUACCAUACAGAAUGGCUAUGCCAAUGAGAUGGACUAUCUGUUUGGCGACGUAAGUAG----------AUCACUAUU----AAAUUGUU (((.(..(((..((..((((.(((((((((((((((((.....)))).))))))).......))))))))))))..).))..).)))..----------.........----........ ( -23.71) >DroVir_CAF1 39628 103 - 1 ACGCACAUUCUGAAAUCGGACUGGUUCUGGUAUACCAUACAGAAUGGCUAUGCCAACGAGAUGGAUUAUCUAUUUGGCGAUGUAAGUGC-------------CAUAUU----AAACACAU ..((.(((((((.....(((....)))(((....)))..)))))))))..((((((..(((((...)))))..))))))......(((.-------------......----...))).. ( -24.70) >DroGri_CAF1 32853 103 - 1 ACGCACAUUCUGAAAUCGGACUGGUUCUGGUAUACAAUCCAGAAUGGCUAUGCCAACGAGAUGGAUUAUCUGUUUGGCGAUGUAAGCAG-------------CAAGAA----ACACCCUU ..((.((.((((....)))).))(((((((........))))))).(((.((((((..(((((...)))))..)))))).....))).)-------------).....----........ ( -26.50) >DroMoj_CAF1 37506 103 - 1 ACUCACAUUCUGAAAUCGGACUGGUUCUGGUAUACCAUACAGAAUGGCUACGCCAACGAGAUGGAUUAUCUAUUUGGCGAUGUAAGUGC-------------AAUCGA----AUACCUAU ..((.(((((((.....(((....)))(((....)))..)))))))((..((((((..(((((...)))))..))))))..))......-------------....))----........ ( -23.60) >DroAna_CAF1 29598 102 - 1 ACUCACAUCCUGAAAUCAGACUGGUUCUGGUAUACCAUCCAGAAUGGCUAUGCCAACGAGAUGGACUAUCUGUUCGGCGACGUAAGUA-----------AUCU---UU----AAACAAUC (((.((.(((((((..((((.(((((((((((((((((.....)))).))))))).......))))))))))))))).)).)).))).-----------....---..----........ ( -27.01) >DroPer_CAF1 28468 120 - 1 ACACACAUCCUGAAAUCAGACUGGUUCUGGUAUACCAUACAGAAUGGCUAUGCCAACGAGAUGGACUAUCUGUUCGGAGACGUAAGUAUCCUUAGCUUUGUCACUGUUCCCUACUUUGUU .........(((((..((((.(((((((((((((((((.....)))).))))))).......))))))))))))))).((((.((((.......)))))))).................. ( -28.41) >consensus ACUCACAUCCUGAAAUCAGACUGGUUCUGGUAUACCAUACAGAAUGGCUAUGCCAACGAGAUGGACUAUCUGUUUGGCGACGUAAGUAC_____________AAUAUU____AAACACUU ....((.(((((((..((((.(((((((((((((((((.....)))).))))))).......))))))))))))))).)).))..................................... (-18.37 = -17.93 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:29 2006