| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,871,108 – 7,871,217 |
| Length | 109 |
| Max. P | 0.894816 |

| Location | 7,871,108 – 7,871,217 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -15.93 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
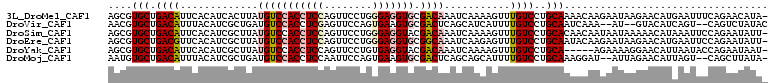
>3L_DroMel_CAF1 7871108 109 + 23771897 AGCGUGCUGACAUUCACAUCACUUAUGUCCACCUCCAGUUCCUGGGAGGUGCGACAAAUCAAAAGUUUGUCCUGCAAAACAAGAAUAAGAACAUGAAUUUCAGAACAUA- ...((.((((.(((((.....(((((((((((((((........))))))).))).........((((........))))....)))))....))))).)))).))...- ( -30.90) >DroVir_CAF1 38094 104 + 1 AACGUGCUGACAUUUACAUCGCUGAUGUCCACCUCGAGUUCCAGUGAAGUGCGACUCAGCAUCAUUUUGUCCUGCAAUCAAA--AU--GUACAUCAGU--CAGUCUAUAC .....((((((.........(((((.((((((.(((........))).))).))))))))..(((((((.........))))--))--).......))--))))...... ( -28.30) >DroSim_CAF1 24360 109 + 1 AGCGUGCUGACAUUCACAUCGCUUAUGUCCACCUCCAGUUCCUGGGAGGUACGACAAAUCAAAAGUUUGUCCUGCACAACAAUAAUAAAAACAUAAAUUCCAGAAUAUU- ...((((.(((((...........))))).((((((........))))))..(((((((.....)))))))..))))................................- ( -23.90) >DroEre_CAF1 27560 109 + 1 AGCGUGCUGACGUUCACAUCGCUUAUGUCCACCUCCAGUUCCUGGGAGGUGCGGCAAAUCAAGAGUUUGUCCUGCAAUACAAGAAUAAGAACAUGAAUUCCAGAAUAUU- ......(((..(((((.....(((((((((((((((........))))))).))).........(((((.....))).))....)))))....)))))..)))......- ( -26.50) >DroYak_CAF1 17219 104 + 1 AGCGUGCUGACAUUCACAUCGCUUAUGUCCACCUCCAGUUCCUGUGAGGUACGACAAAUCAAAAGUUUGUCCUGCA-----AGAAAAGGAACAUUAAUACCAGAAUAAU- ((((((.((.....)))).)))).((((((((((((((...))).)))))..(((((((.....))))))).....-----......)).))))...............- ( -23.50) >DroMoj_CAF1 35977 105 + 1 AAUGUGCUGACAUUUACAUCGCUGAUGUCCACCUCCAAUUCCAGUGAAGUGCGACUCAGCAGCAUUUUGUCCUGCAAAGGAU--AUUAGAACAUUAGU--CAGCUUAUA- .(((.((((((.........(((((.((((((.(((.......).)).))).)))))))).......((((((....)))))--)...........))--)))).))).- ( -32.40) >consensus AGCGUGCUGACAUUCACAUCGCUUAUGUCCACCUCCAGUUCCUGGGAGGUGCGACAAAUCAAAAGUUUGUCCUGCAAAACAAGAAUAAGAACAUGAAUUCCAGAAUAUA_ ....(((.((((.............(((((((((((........))))))).))))...........))))..))).................................. (-15.93 = -16.82 + 0.89)

| Location | 7,871,108 – 7,871,217 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -16.65 |
| Energy contribution | -16.24 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
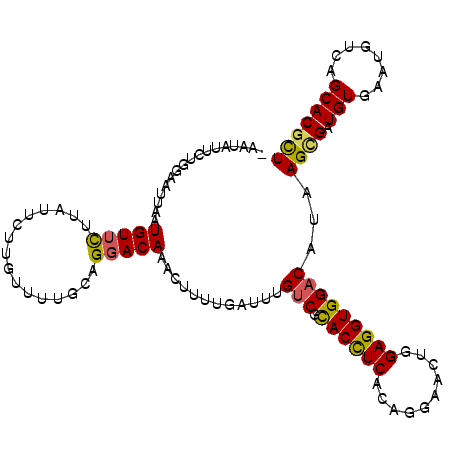
>3L_DroMel_CAF1 7871108 109 - 23771897 -UAUGUUCUGAAAUUCAUGUUCUUAUUCUUGUUUUGCAGGACAAACUUUUGAUUUGUCGCACCUCCCAGGAACUGGAGGUGGACAUAAGUGAUGUGAAUGUCAGCACGCU -...((.((((.(((((((((((((.(((((.....))))).............((((.(((((((........))))))))))))))).))))))))).)))).))... ( -36.90) >DroVir_CAF1 38094 104 - 1 GUAUAGACUG--ACUGAUGUAC--AU--UUUGAUUGCAGGACAAAAUGAUGCUGAGUCGCACUUCACUGGAACUCGAGGUGGACAUCAGCGAUGUAAAUGUCAGCACGUU .....(((((--.(((((((.(--((--((((.........))))))).(((((((((.((((((..........))))))))).))))))......))))))))).))) ( -31.40) >DroSim_CAF1 24360 109 - 1 -AAUAUUCUGGAAUUUAUGUUUUUAUUAUUGUUGUGCAGGACAAACUUUUGAUUUGUCGUACCUCCCAGGAACUGGAGGUGGACAUAAGCGAUGUGAAUGUCAGCACGCU -((((....(((((....)))))...))))...((((..((((.((..(((...((((.(((((((........)))))))))))....))).))...)))).))))... ( -27.40) >DroEre_CAF1 27560 109 - 1 -AAUAUUCUGGAAUUCAUGUUCUUAUUCUUGUAUUGCAGGACAAACUCUUGAUUUGCCGCACCUCCCAGGAACUGGAGGUGGACAUAAGCGAUGUGAACGUCAGCACGCU -......((((..(((((((((((((((..(((...(((((.....)))))...)))..(((((((........))))))))).))))).))))))))..))))...... ( -30.60) >DroYak_CAF1 17219 104 - 1 -AUUAUUCUGGUAUUAAUGUUCCUUUUCU-----UGCAGGACAAACUUUUGAUUUGUCGUACCUCACAGGAACUGGAGGUGGACAUAAGCGAUGUGAAUGUCAGCACGCU -...............((((((.......-----.....(((((((....).))))))..(((((.(((...)))))))))))))).((((.(((........))))))) ( -24.20) >DroMoj_CAF1 35977 105 - 1 -UAUAAGCUG--ACUAAUGUUCUAAU--AUCCUUUGCAGGACAAAAUGCUGCUGAGUCGCACUUCACUGGAAUUGGAGGUGGACAUCAGCGAUGUAAAUGUCAGCACAUU -.....((((--((...((((((...--.........))))))..((..(((((((((.((((((.(.......)))))))))).))))))..))....))))))..... ( -33.70) >consensus _AAUAUUCUGGAAUUAAUGUUCUUAUUCUUGUUUUGCAGGACAAACUUUUGAUUUGUCGCACCUCACAGGAACUGGAGGUGGACAUAAGCGAUGUGAAUGUCAGCACGCU .................(((((................)))))............(((.((((((..........)))))))))...((((.(((........))))))) (-16.65 = -16.24 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:28 2006