
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,858,857 – 7,858,951 |
| Length | 94 |
| Max. P | 0.703422 |

| Location | 7,858,857 – 7,858,951 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.52 |
| Mean single sequence MFE | -29.79 |
| Consensus MFE | -19.71 |
| Energy contribution | -21.22 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7858857 94 + 23771897 AUGGAGACGAACCAAGUUUCAUGGCAAAUGUAUGGCCUGGGAAUUGGCCAU---CAACGCUCCGUCUCUUUCGGGCCAACAAUUGCCGUCUCUUUCG--------- ..(((((((((......)))..(((((.(((.(((((((..(...(((...---.........)))..)..)))))))))).)))))))))))....--------- ( -34.60) >DroSec_CAF1 14579 97 + 1 AUGGAGACGAGCCAAGUUUCAUGGCAUAUGUAUGGCCUGGGAAUUGGCCAUCAUCAACGCUCCGUCUCGUUCGGGCCAACAAUCGCCGUCUCUUUCG--------- ..(((((((.((((.......))))..(((.((((((........)))))))))....((.(((.......)))))..........)))))))....--------- ( -31.40) >DroSim_CAF1 14709 97 + 1 AUGGAGACGAGCCAAGUUUCAUGGCAUAUGUAUGGCCUGGGAAUUGGCCAUCAUCAACGCUCCGUCUCUUUCGGGCCAACAAUCGCCGUCUCUUUCG--------- ..(((((((.((((.......))))...(((.(((((((..(...(((...............)))..)..)))))))))).....)))))))....--------- ( -32.06) >DroEre_CAF1 17802 94 + 1 AUGGAGACGAGCCAAGUUUCAUGGCAAAUGUAUGGCCUGGGAAUUGGCCAUCAU---CGCGCCAUCUCUUUCGGGCCAGCAAUACCCAUCUCUUUCG--------- ..(((((.(.((((.......))))...(((.(((((((..(..(((((.....---.).))))....)..)))))))))).....).)))))....--------- ( -31.40) >DroYak_CAF1 7204 94 + 1 AUGGAGACGAGCCAAGUGCCAUGGCAAAUGUAUGGCCUGGGAAUUGGCCAUCAU---CGCUGCAUCUCUUUCGUGCCAGCACUACCCAUCUCUUUCG--------- ..(((((.((((((.......))))...((.((((((........)))))))))---)(((((((.......))).))))........)))))....--------- ( -27.60) >DroAna_CAF1 17566 92 + 1 AUGGAAGCGAGCCAAGUCUCAU------UGUGGGGCCUGGGAAUUGGCCAUC--------CCCGUCUCUUUUUUUCCGCAUCUUGCUGUCUCUGUCGCUGCAUACA (((..(((((.(..((.(.((.------(((((((...((((..(((.....--------.)))..))))...)))))))...))..).))..)))))).)))... ( -21.70) >consensus AUGGAGACGAGCCAAGUUUCAUGGCAAAUGUAUGGCCUGGGAAUUGGCCAUCAU___CGCUCCGUCUCUUUCGGGCCAACAAUAGCCGUCUCUUUCG_________ ..(((((((.(((((.(((((.(((.........)))))))).)))))..........((.(((.......)))))..........)))))))............. (-19.71 = -21.22 + 1.50)

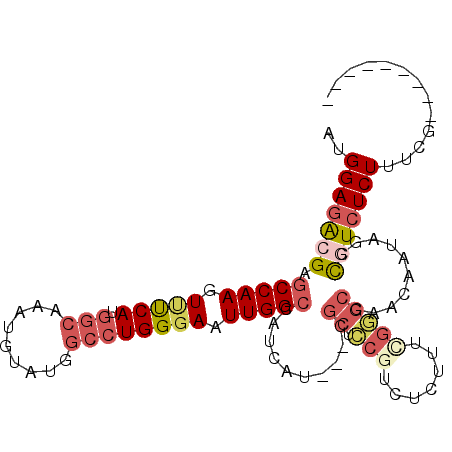

| Location | 7,858,857 – 7,858,951 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.52 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -19.29 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7858857 94 - 23771897 ---------CGAAAGAGACGGCAAUUGUUGGCCCGAAAGAGACGGAGCGUUG---AUGGCCAAUUCCCAGGCCAUACAUUUGCCAUGAAACUUGGUUCGUCUCCAU ---------.....((((((((((.(((((((((....)....((.(.((((---.....)))).))).))))).))).)))))..(((......))))))))... ( -32.80) >DroSec_CAF1 14579 97 - 1 ---------CGAAAGAGACGGCGAUUGUUGGCCCGAACGAGACGGAGCGUUGAUGAUGGCCAAUUCCCAGGCCAUACAUAUGCCAUGAAACUUGGCUCGUCUCCAU ---------(....)....(((.(....).))).....(((((((.......(((((((((........)))))).)))..((((.......)))))))))))... ( -31.60) >DroSim_CAF1 14709 97 - 1 ---------CGAAAGAGACGGCGAUUGUUGGCCCGAAAGAGACGGAGCGUUGAUGAUGGCCAAUUCCCAGGCCAUACAUAUGCCAUGAAACUUGGCUCGUCUCCAU ---------(....)....(((.(....).))).....(((((((.......(((((((((........)))))).)))..((((.......)))))))))))... ( -31.50) >DroEre_CAF1 17802 94 - 1 ---------CGAAAGAGAUGGGUAUUGCUGGCCCGAAAGAGAUGGCGCG---AUGAUGGCCAAUUCCCAGGCCAUACAUUUGCCAUGAAACUUGGCUCGUCUCCAU ---------.....(((((((((..........(....)..((((((.(---(((((((((........)))))).))))))))))........)))))))))... ( -34.60) >DroYak_CAF1 7204 94 - 1 ---------CGAAAGAGAUGGGUAGUGCUGGCACGAAAGAGAUGCAGCG---AUGAUGGCCAAUUCCCAGGCCAUACAUUUGCCAUGGCACUUGGCUCGUCUCCAU ---------.....((((((((((((((((((((....).........(---(((((((((........)))))).))))))))..))))))..)))))))))... ( -39.30) >DroAna_CAF1 17566 92 - 1 UGUAUGCAGCGACAGAGACAGCAAGAUGCGGAAAAAAAGAGACGGG--------GAUGGCCAAUUCCCAGGCCCCACA------AUGAGACUUGGCUCGCUUCCAU ...(((.(((((..(((...((.....))..............(((--------(.(((.......)))..))))...------......)))...)))))..))) ( -20.70) >consensus _________CGAAAGAGACGGCAAUUGUUGGCCCGAAAGAGACGGAGCG___AUGAUGGCCAAUUCCCAGGCCAUACAUUUGCCAUGAAACUUGGCUCGUCUCCAU .........(....).......................(((((((..........((((((........))))))......((((.......)))))))))))... (-19.29 = -19.98 + 0.70)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:24 2006