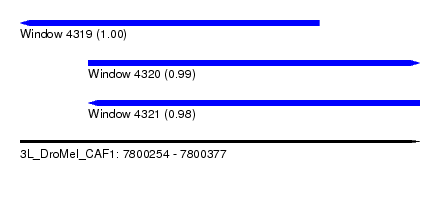
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,800,254 – 7,800,377 |
| Length | 123 |
| Max. P | 0.995592 |

| Location | 7,800,254 – 7,800,346 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -19.72 |
| Energy contribution | -19.48 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7800254 92 - 23771897 CCUUGCCACCGCUGAGCAGCUUGCCACUGCAAGUGGUGCACAAUCUGCCGCAUUUGUUGGCCGGCAACAGUGGCGGCAACACGGGCAACAAU--- ..(((((.((((((....(((.((((..((((((((.(((.....)))).))))))))))).)))..)))))).)))))....(....)...--- ( -37.70) >DroSec_CAF1 9952 92 - 1 CCUUGCCACCGCUGAGCAGCUUGCCACUGCAAGUGGUGCACAAUCUGCCGCACUUGUUGGCCGGCAACAGUGGCGGCAACAAUGCCAACAAC--- ..(((((.((((((....(((.((((..((((((((.(((.....)))).))))))))))).)))..)))))).))))).............--- ( -39.00) >DroWil_CAF1 38579 86 - 1 CAUUGCCGCCCUUGAGUAGUUUACCUCUGCAAGUGGUUAAUAAUUUGCCACAUUUAUUGG---GCAAUAAUAACAAUAAU---AAUAACAAU--- .......((((....((((.......))))..(((((.........))))).......))---))...............---.........--- ( -18.10) >DroYak_CAF1 10521 95 - 1 CCUUGCCGCCGCUGAGCAGCUUGCCACUGCAAGUGGUGCACAAUCUGCCGCAUUUGUUGGCCGGCAACGGUGGCAGCAACGCGGGCAACAAUGCC ..((((.(((((((....(((.((((..((((((((.(((.....)))).))))))))))).)))..))))))).))))....((((....)))) ( -38.80) >DroMoj_CAF1 27467 77 - 1 CAUUGCCACCAUUGAGCAGCCUGCCGCUGCAGGUGGUCCACAAUUUGCCCCAUUUGCUGGCUGGCAA---------------CACGAACAAC--- ...(((((....)).)))(((.((((..((((((((..((.....))..)))))))))))).)))..---------------..........--- ( -25.30) >consensus CCUUGCCACCGCUGAGCAGCUUGCCACUGCAAGUGGUGCACAAUCUGCCGCAUUUGUUGGCCGGCAACAGUGGCAGCAAC__GGCCAACAAU___ ..(((((((((((..((((.......)))).)))))).........((((.......)))).)))))............................ (-19.72 = -19.48 + -0.24)

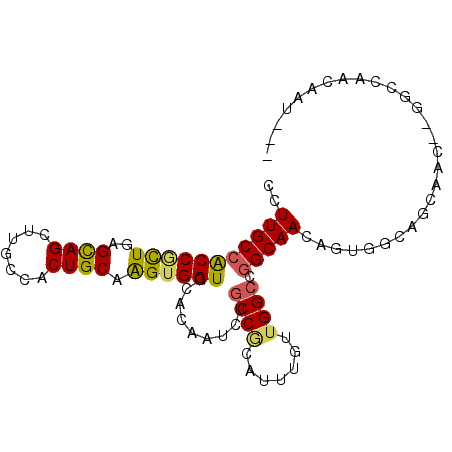
| Location | 7,800,275 – 7,800,377 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -43.98 |
| Consensus MFE | -25.20 |
| Energy contribution | -24.10 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7800275 102 + 23771897 A------CUGUUGCCGGCCAACAAAUGCGGCAGAUUGUGCACCACUUGCAGUGGCAAGCUGCUCAGCGGUGGCAAGG------CUGCCGUCUGCGACAUGUUGCCGAGCAACUG .------..(((((((((.((((.....(((((.((((.((((.((.(((((.....)))))..)).))))))))..------)))))(((...))).)))))))).))))).. ( -46.60) >DroVir_CAF1 28115 108 + 1 GUUGUUGCCAUUGCCUGCCAGCAGGUGUGGCAAAUUGUGCACCACCUGCAGCGGCAGGCUGCUCAGUGGCGGCAACG------UCGCCGCUUGCGACAUAUUGCCAAGCAUUUG (((((((((((((((((((.(((((((..(((.....)))..)))))))...))))))).....))))))))))))(------((((.....)))))....(((...))).... ( -55.40) >DroSec_CAF1 9973 102 + 1 A------CUGUUGCCGGCCAACAAGUGCGGCAGAUUGUGCACCACUUGCAGUGGCAAGCUGCUCAGCGGUGGCAAGG------CCGCCGUCUGCGACAUGUUGCCGAGCAACUG .------..(((((((((.((((.(((((((...((((.((((.((.(((((.....)))))..)).)))))))).)------))))((....)))).)))))))).))))).. ( -47.60) >DroWil_CAF1 38597 105 + 1 A------UUAUUGC---CCAAUAAAUGUGGCAAAUUAUUAACCACUUGCAGAGGUAAACUACUCAAGGGCGGCAAUGUUGCUGCCGUCGUUUGCGACAUGUUGCCCAACAGUUG .------...((((---(((.....)).))))).....((((........(((........)))..((((((((.((((((.((....))..))))))))))))))....)))) ( -31.90) >DroYak_CAF1 10545 102 + 1 A------CCGUUGCCGGCCAACAAAUGCGGCAGAUUGUGCACCACUUGCAGUGGCAAGCUGCUCAGCGGCGGCAAGG------CUGCCGUCUGCGACAUGUUGCCGAGCAACUG .------..(((((((((.((((..((..((((((((.(((...(((((....))))).))).)))(((((((...)------)))))))))))..)))))))))).))))).. ( -46.10) >DroMoj_CAF1 27477 98 + 1 ----------UUGCCAGCCAGCAAAUGGGGCAAAUUGUGGACCACCUGCAGCGGCAGGCUGCUCAAUGGUGGCAAUG------UUGUUCCUUGCGACAUGUUGCCCAGCAUUUG ----------...........((((((((((((......(.(((((.(((((.....))))).....)))))).(((------((((.....))))))).))))))..)))))) ( -36.30) >consensus A______CUGUUGCCGGCCAACAAAUGCGGCAAAUUGUGCACCACUUGCAGUGGCAAGCUGCUCAGCGGCGGCAAGG______CCGCCGUCUGCGACAUGUUGCCGAGCAACUG .........(((((..(.((((..(((..((((..((.(((...(((((....))))).))).))(((((((...........)))))))))))..))))))).)..))))).. (-25.20 = -24.10 + -1.10)
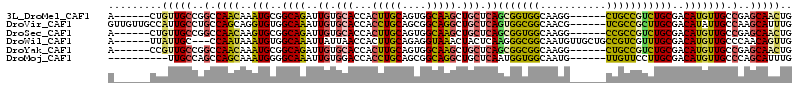

| Location | 7,800,275 – 7,800,377 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -43.13 |
| Consensus MFE | -32.67 |
| Energy contribution | -31.73 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7800275 102 - 23771897 CAGUUGCUCGGCAACAUGUCGCAGACGGCAG------CCUUGCCACCGCUGAGCAGCUUGCCACUGCAAGUGGUGCACAAUCUGCCGCAUUUGUUGGCCGGCAACAG------U ..(((((.(((((((((((.(((((.(((..------....)))((((((..((((.......)))).))))))......))))).)))..)))).)))))))))..------. ( -46.10) >DroVir_CAF1 28115 108 - 1 CAAAUGCUUGGCAAUAUGUCGCAAGCGGCGA------CGUUGCCGCCACUGAGCAGCCUGCCGCUGCAGGUGGUGCACAAUUUGCCACACCUGCUGGCAGGCAAUGGCAACAAC ....((((((((...(((((((.....))))------)))....))))...))))((((((((..(((((((..(((.....)))..)))))))))))))))..((....)).. ( -53.60) >DroSec_CAF1 9973 102 - 1 CAGUUGCUCGGCAACAUGUCGCAGACGGCGG------CCUUGCCACCGCUGAGCAGCUUGCCACUGCAAGUGGUGCACAAUCUGCCGCACUUGUUGGCCGGCAACAG------U ..(((((.(((((((((((.(((((((((((------........)))))).(((((((((....))))))..)))....))))).)))..)))).)))))))))..------. ( -47.50) >DroWil_CAF1 38597 105 - 1 CAACUGUUGGGCAACAUGUCGCAAACGACGGCAGCAACAUUGCCGCCCUUGAGUAGUUUACCUCUGCAAGUGGUUAAUAAUUUGCCACAUUUAUUGG---GCAAUAA------U .(((..((((((.....((((....))))(((((.....)))))))))....((((.......)))).))..)))......(((((.((.....)))---))))...------. ( -32.10) >DroYak_CAF1 10545 102 - 1 CAGUUGCUCGGCAACAUGUCGCAGACGGCAG------CCUUGCCGCCGCUGAGCAGCUUGCCACUGCAAGUGGUGCACAAUCUGCCGCAUUUGUUGGCCGGCAACGG------U ..(((((.(((((((((((.(((((.(((..------....)))((((((..((((.......)))).))))))......))))).)))..)))).)))))))))..------. ( -46.40) >DroMoj_CAF1 27477 98 - 1 CAAAUGCUGGGCAACAUGUCGCAAGGAACAA------CAUUGCCACCAUUGAGCAGCCUGCCGCUGCAGGUGGUCCACAAUUUGCCCCAUUUGCUGGCUGGCAA---------- ..((((...(((((..((((....)..))).------..)))))..)))).....(((.((((..((((((((..((.....))..)))))))))))).)))..---------- ( -33.10) >consensus CAAUUGCUCGGCAACAUGUCGCAAACGGCAG______CCUUGCCACCGCUGAGCAGCUUGCCACUGCAAGUGGUGCACAAUCUGCCGCAUUUGUUGGCCGGCAACAG______U ....((((.(.((((((((.(((((.(((((........)))))((((((..((((.......)))).))))))......))))).)))..))))).).))))........... (-32.67 = -31.73 + -0.94)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:10 2006