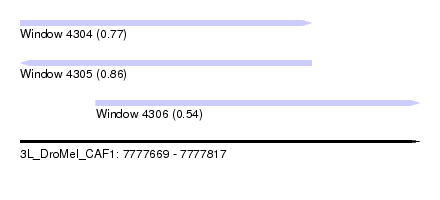
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,777,669 – 7,777,817 |
| Length | 148 |
| Max. P | 0.860569 |

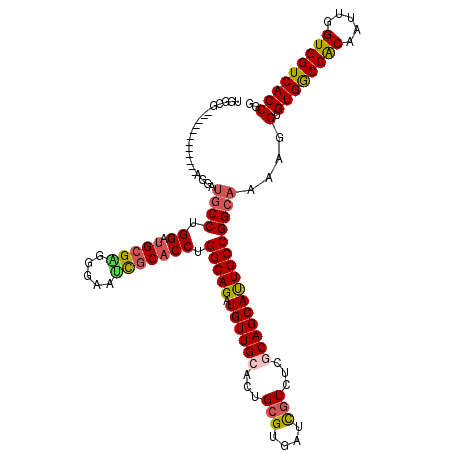
| Location | 7,777,669 – 7,777,777 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -48.28 |
| Consensus MFE | -39.52 |
| Energy contribution | -39.76 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7777669 108 + 23771897 UGGCG------------AGGAUGCCUGGAUGCGAGGGAAUCGCACCUGGCAGAUGUUGCACUGCGUGAUCGUCUCACAGCAUUGCCGGCAAAAGGUGUGACCACAAUUGGUGGUCACCGG ((.((------------....((((.((.(((((.....))))))).))))......(((.(((((((.....)))).))).))))).))....(.((((((((.....))))))))).. ( -43.80) >DroPse_CAF1 21156 120 + 1 GCCUCCGCCGCGCGGGGAGGCGUCCUGGGCGCGGGGGAACUUCACCUGGCAGAUGUUGCACUGGGUGAUCGUCUCGCAGCACUGCCGGCAGAAGGUGUGGCCGCAGUUGGUGGUCACCGG .(((((((.((.(((((.....))))).)))))))))..((((..(((((((.((((((...(((.......))))))))))))))))..))))(.((((((((.....))))))))).. ( -61.50) >DroSec_CAF1 18052 108 + 1 UGGCG------------AGGAUGCCUGGAUGCGAGGGAAUCGCACCUGGCAGAUGUUGCACUGCGUGAUCGUCUCACAGCAUUGCCGGCAAAAGGUGUGACCACAAUUGGUGGUCACCGG ((.((------------....((((.((.(((((.....))))))).))))......(((.(((((((.....)))).))).))))).))....(.((((((((.....))))))))).. ( -43.80) >DroEre_CAF1 18094 108 + 1 UGGCG------------AGGAUGCCUGGAUGCGUGGGAAGCGCACCUGGCAGAUGUUGCACUGCGUGAUCGUCUCGCAGCACUGCCGGCAAAAGGUGUGGCCACAAUUGGUGGUCACCGG ...((------------....((((.((.(((((.....))))))).(((((.((((((...(((....)))...)))))))))))))))......((((((((.....)))))))))). ( -47.80) >DroYak_CAF1 19280 108 + 1 UGGCG------------AGGAUGCCUGGAUGCGGGGGAAACGCACCUGGCAGAUGUUGCACUGCGUGAUUGUUUCGCAGCAUUGCCGGCAAAAGGUGUGGCCACAAUUGGUGGUCACCGG .(((.------------....((((.((.((((.......)))))).))))((((((((...(((....)))...)))))))))))........(.((((((((.....))))))))).. ( -44.50) >consensus UGGCG____________AGGAUGCCUGGAUGCGAGGGAAUCGCACCUGGCAGAUGUUGCACUGCGUGAUCGUCUCGCAGCAUUGCCGGCAAAAGGUGUGGCCACAAUUGGUGGUCACCGG .....................((((.((.(((((.....))))))).(((((.((((((...(((....)))...)))))))))))))))....(.((((((((.....))))))))).. (-39.52 = -39.76 + 0.24)

| Location | 7,777,669 – 7,777,777 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -30.20 |
| Energy contribution | -31.04 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7777669 108 - 23771897 CCGGUGACCACCAAUUGUGGUCACACCUUUUGCCGGCAAUGCUGUGAGACGAUCACGCAGUGCAACAUCUGCCAGGUGCGAUUCCCUCGCAUCCAGGCAUCCU------------CGCCA ...((((((((.....))))))))..........(((...((((((.(.....).))))))........((((.(((((((.....)))))))..))))....------------.))). ( -41.40) >DroPse_CAF1 21156 120 - 1 CCGGUGACCACCAACUGCGGCCACACCUUCUGCCGGCAGUGCUGCGAGACGAUCACCCAGUGCAACAUCUGCCAGGUGAAGUUCCCCCGCGCCCAGGACGCCUCCCCGCGCGGCGGAGGC ..(((....))).......(((....((((.((((((((((.((((.(........)...)))).)).))))).))))))).(((.((((((...(((....)))..)))))).)))))) ( -44.80) >DroSec_CAF1 18052 108 - 1 CCGGUGACCACCAAUUGUGGUCACACCUUUUGCCGGCAAUGCUGUGAGACGAUCACGCAGUGCAACAUCUGCCAGGUGCGAUUCCCUCGCAUCCAGGCAUCCU------------CGCCA ...((((((((.....))))))))..........(((...((((((.(.....).))))))........((((.(((((((.....)))))))..))))....------------.))). ( -41.40) >DroEre_CAF1 18094 108 - 1 CCGGUGACCACCAAUUGUGGCCACACCUUUUGCCGGCAGUGCUGCGAGACGAUCACGCAGUGCAACAUCUGCCAGGUGCGCUUCCCACGCAUCCAGGCAUCCU------------CGCCA ..((((.((((.....))))...)))).......(((.(..(((((.(.....).)))))..)......((((.((((((.......))))))..))))....------------.))). ( -41.10) >DroYak_CAF1 19280 108 - 1 CCGGUGACCACCAAUUGUGGCCACACCUUUUGCCGGCAAUGCUGCGAAACAAUCACGCAGUGCAACAUCUGCCAGGUGCGUUUCCCCCGCAUCCAGGCAUCCU------------CGCCA ..(((((((((.....))))..........((((((((((((((((.........))))).....))).)))).((((((.......))))))..))))...)------------)))). ( -35.90) >consensus CCGGUGACCACCAAUUGUGGCCACACCUUUUGCCGGCAAUGCUGCGAGACGAUCACGCAGUGCAACAUCUGCCAGGUGCGAUUCCCCCGCAUCCAGGCAUCCU____________CGCCA ..((((.((((.....))))...))))...(((((((((((.((((...((....))...)))).))).)))).(((((((.....)))))))..))))..................... (-30.20 = -31.04 + 0.84)

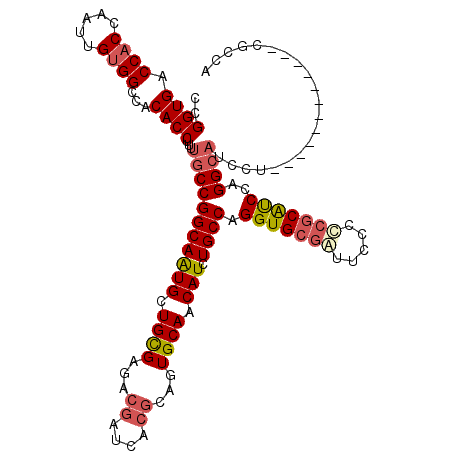
| Location | 7,777,697 – 7,777,817 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -47.34 |
| Consensus MFE | -40.26 |
| Energy contribution | -39.98 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7777697 120 + 23771897 CGCACCUGGCAGAUGUUGCACUGCGUGAUCGUCUCACAGCAUUGCCGGCAAAAGGUGUGACCACAAUUGGUGGUCACCGGACAGCGCAGGAUAUCGCUGCAAAGCGGACACAUUAGCGGA (((.((((((...(((((((.(((((((.....)))).))).)).)))))....(.((((((((.....))))))))).....)).))))...(((((....)))))........))).. ( -45.60) >DroPse_CAF1 21196 120 + 1 UUCACCUGGCAGAUGUUGCACUGGGUGAUCGUCUCGCAGCACUGCCGGCAGAAGGUGUGGCCGCAGUUGGUGGUCACCGGACAGCGGAGGAUGUCCCUGCAGAGCGGACAGAGCAGCGGG .((((((.((((...))))...))))))....(((((.((.((((((((((...(.((((((((.....)))))))))(((((........)))))))))....))).))).)).))))) ( -50.80) >DroSec_CAF1 18080 120 + 1 CGCACCUGGCAGAUGUUGCACUGCGUGAUCGUCUCACAGCAUUGCCGGCAAAAGGUGUGACCACAAUUGGUGGUCACCGGACAGCGCAGGAUAUCGCCGCAAAGCGGACACAUCAGCGGA (((.((((((...(((((((.(((((((.....)))).))).)).)))))....(.((((((((.....))))))))).....)).)))).....(((((...)))).)......))).. ( -44.80) >DroEre_CAF1 18122 120 + 1 CGCACCUGGCAGAUGUUGCACUGCGUGAUCGUCUCGCAGCACUGCCGGCAAAAGGUGUGGCCACAAUUGGUGGUCACCGGACAGCGCAGGAUAUCGCCGCACAGCGGACACAUCAGCGGA (((..(((((((.((((((...(((....)))...))))))))))))).....(((((((((((.....)))))..(((....((((........).)))....))).)))))).))).. ( -47.30) >DroYak_CAF1 19308 120 + 1 CGCACCUGGCAGAUGUUGCACUGCGUGAUUGUUUCGCAGCAUUGCCGGCAAAAGGUGUGGCCACAAUUGGUGGUCACCGGACAGCGCAGGAUGUCGCCGCACAGCGGACACAUCAGCGGA .((..(((((((.((((((...(((....)))...)))))))))))))......(.((((((((.....))))))))).....))((..(((((..((((...))))..))))).))... ( -48.20) >consensus CGCACCUGGCAGAUGUUGCACUGCGUGAUCGUCUCGCAGCAUUGCCGGCAAAAGGUGUGGCCACAAUUGGUGGUCACCGGACAGCGCAGGAUAUCGCCGCAAAGCGGACACAUCAGCGGA (((..(((((((.((((((...(((....)))...)))))))))))))......((((((((((.....)))))).(((....((((........)).))....)))))))....))).. (-40.26 = -39.98 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:56 2006