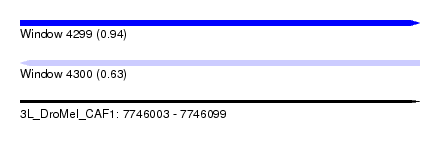
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,746,003 – 7,746,099 |
| Length | 96 |
| Max. P | 0.936398 |

| Location | 7,746,003 – 7,746,099 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 59.41 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -10.08 |
| Energy contribution | -9.97 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
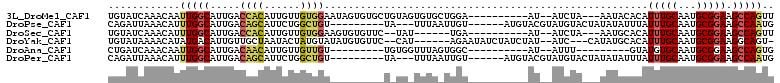
>3L_DroMel_CAF1 7746003 96 + 23771897 UGUAUCAAACAAUUGGCAUUGACCACAUUGUUGUGGAAUAGUGUGCUGUAGUGUGCUGGA----------AU--AUCUA---AAUACACAUUUGCAAUGCGGAAGCCAGUU ..........(((((((.....(((((....)))))....((((((...((((((.(((.----------..--..)))---....)))))).)).))))....))))))) ( -24.80) >DroPse_CAF1 19925 93 + 1 CAGAUUAAACAUUUGGCAUUGACAGCAUUCUGGCUGU---------UA---UUUAAUUGU------AUGUACGUAUGUACUAUAUAUUUAUUUGCAAUGCGGAAGCCAAUG ............(((((..(((((((......)))))---------))---(((.(((((------(.(((.((((((...)))))).))).))))))...)))))))).. ( -23.50) >DroSec_CAF1 10530 88 + 1 UGUAUCAAACAUUUGGCAUUGACCACAUUGUUGUGGAAGUGUGUUC--UAU------UGA----------AU--AUCUA---AAUGCACAUUUGCAAUGCGGAAGCCAGUU ...........(((.((((((.(((((....)))))((((((((..--...------.(.----------..--..)..---...)))))))).)))))).)))....... ( -23.40) >DroYak_CAF1 11115 97 + 1 UGUAUAAAACAUAUGACAUUGUUGCUAAUACUAUGUAUAUGUGUUC--CAU------AGAAUAUCUAUCUAU--AUC---CAUAUGCACAUUUGCAAUGCGGAAGGCAGU- .((((..((((........))))....))))........(((.(((--(((------(((.......)))))--...---(((.((((....))))))).)))).)))..- ( -17.80) >DroAna_CAF1 17815 81 + 1 CUGAUCAAACAAUUGGCAUUGACAACAUUGUUGUUGU---------UGUGGUUUAGUGGC----------AU--AUUU---------GUAUGUGCAAUGCGGAAGCCAGUG ...........((((((...(((((((....))))))---------)........((.((----------((--((..---------..))))))...))....)))))). ( -21.60) >DroPer_CAF1 19982 93 + 1 CAGAUUAAACAUUUGGCAUUGACAGCAUUCUGGCUGU---------UA---UUUAAUUGU------AUGUACGUAUGUACUAUAUAUUUAUUUGCAAUGCGGAAGCCAAUG ............(((((..(((((((......)))))---------))---(((.(((((------(.(((.((((((...)))))).))).))))))...)))))))).. ( -23.50) >consensus CGGAUCAAACAUUUGGCAUUGACAACAUUGUUGUGGA_________U_UA_UUUA_UUGA__________AU__AUCUA___UAUACACAUUUGCAAUGCGGAAGCCAGUG ............(((((.....((((......))))......................................................(((((...))))).))))).. (-10.08 = -9.97 + -0.11)
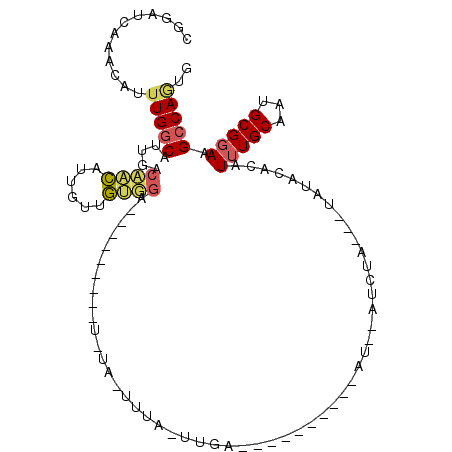
| Location | 7,746,003 – 7,746,099 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 59.41 |
| Mean single sequence MFE | -15.90 |
| Consensus MFE | -7.14 |
| Energy contribution | -6.87 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7746003 96 - 23771897 AACUGGCUUCCGCAUUGCAAAUGUGUAUU---UAGAU--AU----------UCCAGCACACUACAGCACACUAUUCCACAACAAUGUGGUCAAUGCCAAUUGUUUGAUACA ...((((..((((((((....((((....---..(..--..----------..).((........)).........)))).)))))))).....))))............. ( -18.20) >DroPse_CAF1 19925 93 - 1 CAUUGGCUUCCGCAUUGCAAAUAAAUAUAUAGUACAUACGUACAU------ACAAUUAAA---UA---------ACAGCCAGAAUGCUGUCAAUGCCAAAUGUUUAAUCUG ..(((((........................((((....))))..------.........---..---------(((((......)))))....)))))............ ( -15.50) >DroSec_CAF1 10530 88 - 1 AACUGGCUUCCGCAUUGCAAAUGUGCAUU---UAGAU--AU----------UCA------AUA--GAACACACUUCCACAACAAUGUGGUCAAUGCCAAAUGUUUGAUACA ...((((..((((((((....((((....---.....--.(----------((.------...--)))........)))).)))))))).....))))............. ( -17.07) >DroYak_CAF1 11115 97 - 1 -ACUGCCUUCCGCAUUGCAAAUGUGCAUAUG---GAU--AUAGAUAGAUAUUCU------AUG--GAACACAUAUACAUAGUAUUAGCAACAAUGUCAUAUGUUUUAUACA -.......((((...((((....))))..))---))(--(((((.......)))------)))--....((((((((((.((.......)).)))).))))))........ ( -16.70) >DroAna_CAF1 17815 81 - 1 CACUGGCUUCCGCAUUGCACAUAC---------AAAU--AU----------GCCACUAAACCACA---------ACAACAACAAUGUUGUCAAUGCCAAUUGUUUGAUCAG ..((((.....((((((..((((.---------...)--))----------).............---------((((((....))))))))))))(((....))).)))) ( -12.40) >DroPer_CAF1 19982 93 - 1 CAUUGGCUUCCGCAUUGCAAAUAAAUAUAUAGUACAUACGUACAU------ACAAUUAAA---UA---------ACAGCCAGAAUGCUGUCAAUGCCAAAUGUUUAAUCUG ..(((((........................((((....))))..------.........---..---------(((((......)))))....)))))............ ( -15.50) >consensus CACUGGCUUCCGCAUUGCAAAUAUGUAUA___UAGAU__AU__________UCAA_UAAA_UA_A_________ACAACAACAAUGUUGUCAAUGCCAAAUGUUUGAUACA ...((((....((...)).........................................................((((......)))).....))))............. ( -7.14 = -6.87 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:50 2006