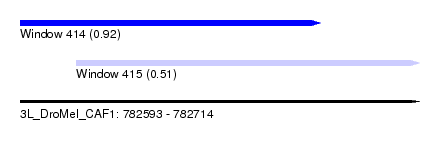
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 782,593 – 782,714 |
| Length | 121 |
| Max. P | 0.915527 |

| Location | 782,593 – 782,684 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -23.91 |
| Consensus MFE | -15.11 |
| Energy contribution | -16.47 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
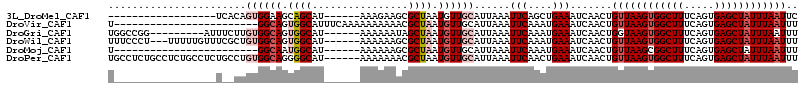
>3L_DroMel_CAF1 782593 91 + 23771897 ------------------UCACAGUGGAAGCAGCAU------AAAGAAGCGCUAAUGUUGCAUUAAAUUCAGCUGAAAUCAACUGUUAAGUGGCUUUCAGUGAGCUAUUUAAUUC ------------------..(((((.((.(((((((------..((.....)).)))))))......(((....))).)).)))))((((((((((.....)))))))))).... ( -23.30) >DroVir_CAF1 163401 91 + 1 U------------------------GGCAGUGGCAUUUCAAAAAAAAAACGCUAAUGUUGCAUUAAAUUCAAAUGAAAUCAACUGUUAAGUGGCUUUCAGUGAGCUAUUUAAUUU .------------------------.(((((.(.((((((..........((.......))............))))))).)))))((((((((((.....)))))))))).... ( -18.05) >DroGri_CAF1 182127 100 + 1 UGGCCGG---------AUUUCUUGUGGCAGUGGCAU------AAAAAAUAGCUAAUGUUGCAUUAAAUUCAAAUGAAAUCAACUGGUAAGUGGCUUUCAGUGAGCUAUUUAAUUU ..(((((---------((((..((..(((.((((..------........)))).)))..))..))))))...((....))...)))(((((((((.....)))))))))..... ( -27.80) >DroWil_CAF1 159541 106 + 1 UUUCCCU---UUUUUGUUUCGCUGUGGCAGUGGCAU------AAAAAAGCGCUAAUGUUGCAUUAAAUUCAAAUGAAAUCAACUGUUAAGUGGCUUUCAGUGAGCUAUUUAAUUU ((((.((---(((((((.((((((...)))))).))------))))))).((.......)).............))))......((((((((((((.....)))))))))))).. ( -25.30) >DroMoj_CAF1 159978 85 + 1 U------------------------GGCAAUGGCAU------AAAAAAGCGCUAAUGUUGCAUUAAAUUCAAAUGAAAUCAACUGUUAAGCGGCUUUCAGUGAGCUAUUUAAUUU .------------------------...((((((.(------(..(((((((((((((((((((.......))))....)))).))).))).)))))...)).))))))...... ( -14.80) >DroPer_CAF1 158459 109 + 1 UGCCUCUGCCUCUGCCUCUGCCUGUGGCAGGGGCAU------AAAAAAACGCUAAUGUUGCAUUAAAUUCAACUGAAAUCAACUGUUAAGUGGCUUUCAGUGAGCUAUUUAAUUU ((((((((((.(.((....))..).)))))))))).------........((.......))(((((((...(((((((.(.(((....))).).))))))).....))))))).. ( -34.20) >consensus U_________________U__CUGUGGCAGUGGCAU______AAAAAAGCGCUAAUGUUGCAUUAAAUUCAAAUGAAAUCAACUGUUAAGUGGCUUUCAGUGAGCUAUUUAAUUU .......................((((((.((((................)))).))))))......(((....))).......((((((((((((.....)))))))))))).. (-15.11 = -16.47 + 1.36)
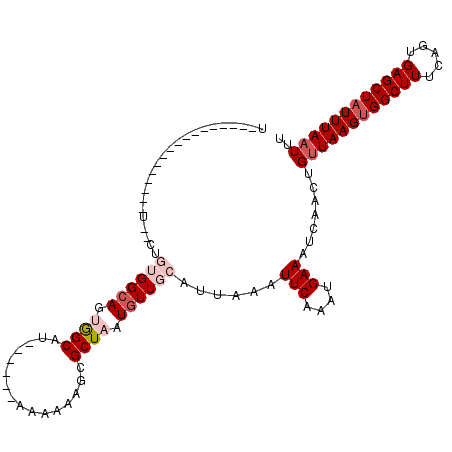
| Location | 782,610 – 782,714 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -18.06 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.85 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 782610 104 + 23771897 U------AAAGAAGCGCUAAUGUUGCAUUAAAUUCAGCUGAAAUCAACUGUUAAGUGGCUUUCAGUGAGCUAUUUAAUUCUCGAACACCC-----AAAAAGAGCAAAGGC-GAUUC---- .------...(((.((((....((((........(((.((....)).)))(((((((((((.....))))))))))).............-----.......)))).)))-).)))---- ( -23.30) >DroVir_CAF1 163412 108 + 1 UUUCAAAAAAAAAACGCUAAUGUUGCAUUAAAUUCAAAUGAAAUCAACUGUUAAGUGGCUUUCAGUGAGCUAUUUAAUUUUCCAACACCA-----AAAAAGAGCUAAGAA-AAU------ ...............(((..(((((.......(((....))).......((((((((((((.....))))))))))))....)))))...-----......)))......-...------ ( -16.00) >DroPse_CAF1 152622 107 + 1 U------AAAAAAACGCUAAUGUUGCAUUAAAUUCAACUGAAAUCAACUGUUAAGUGGCUUUCAGUGAGCUAUUUAAUUUCAAAACAUCC-----AAAAAGAGCAAAUAA-AAA-UAAAC .------........(((....(((.......(((....))).......((((((((((((.....))))))))))))...........)-----))....)))......-...-..... ( -16.60) >DroGri_CAF1 182153 103 + 1 U------AAAAAAUAGCUAAUGUUGCAUUAAAUUCAAAUGAAAUCAACUGGUAAGUGGCUUUCAGUGAGCUAUUUAAUUUUCCAACACCAA----AAAAAGAGCUAAGCA-AAU------ .------......(((((...((((((((.......))))....))))(((((((((((((.....)))))))))).....))).......----......)))))....-...------ ( -17.60) >DroMoj_CAF1 159989 108 + 1 U------AAAAAAGCGCUAAUGUUGCAUUAAAUUCAAAUGAAAUCAACUGUUAAGCGGCUUUCAGUGAGCUAUUUAAUUUCCCUACGCCGAGCUGAAAAAGAGCUGCGAAAAAU------ .------.....(((((.......))......(((....))).......)))..((((((((((((..((................))...)))))....))))))).......------ ( -18.29) >DroPer_CAF1 158494 107 + 1 U------AAAAAAACGCUAAUGUUGCAUUAAAUUCAACUGAAAUCAACUGUUAAGUGGCUUUCAGUGAGCUAUUUAAUUUCAAAACAUCC-----AAAAAGAGCAAAUAA-AAA-UAUAC .------........(((....(((.......(((....))).......((((((((((((.....))))))))))))...........)-----))....)))......-...-..... ( -16.60) >consensus U______AAAAAAACGCUAAUGUUGCAUUAAAUUCAAAUGAAAUCAACUGUUAAGUGGCUUUCAGUGAGCUAUUUAAUUUCCCAACACCC_____AAAAAGAGCAAAGAA_AAU______ ...............(((...((((.(((....((....))))))))).((((((((((((.....)))))))))))).......................)))................ (-14.52 = -14.85 + 0.33)

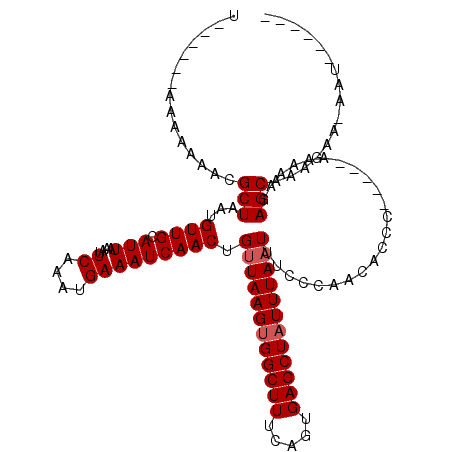
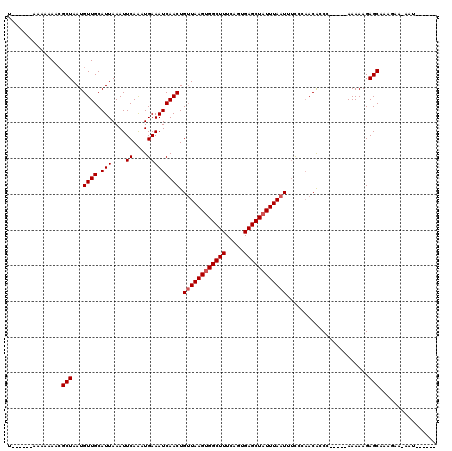
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:06 2006