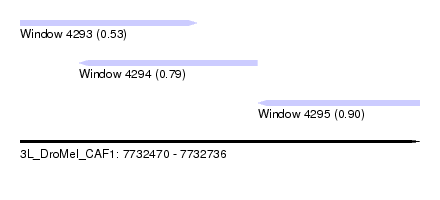
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,732,470 – 7,732,736 |
| Length | 266 |
| Max. P | 0.897391 |

| Location | 7,732,470 – 7,732,588 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.30 |
| Mean single sequence MFE | -39.16 |
| Consensus MFE | -26.52 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7732470 118 + 23771897 CAAUAUUACCUGCAACGCGG-GUUAUAUCAUGCAGGGCUCACAAUUUGCGGUUUGUGGACACACCGGACUUUGGUCUAGUCCAGAAGCCAAGUGCGUGGGUAAGUACACAUCAAUGU-AC ...(((.((((((...))))-)).)))(((((((..((.........))(((((.(((((..((((.....))))...))))).)))))...)))))))....(((((......)))-)) ( -37.50) >DroSec_CAF1 28906 118 + 1 CAAUAUUACCUGCAACGCGGGGUUAUAUUCUGCAGGGUUCACAAUUCGCGAGUUGUGGACACACCGGUCUUUGGUCCAGUCCAGAAGCCAAGUGCGUGGGUAAGUACAAAUCAAUGA-A- .....(((((..(.(((((((((...))))))).((((((((((((....))))))))))...))((..(((((......)))))..))..))..)..)))))..............-.- ( -38.30) >DroSim_CAF1 25035 117 + 1 CAAUAUUACCUGCAACGCGG-GUUAUAUCCUGCAGGGUUCACAAUUCGCGAGUUGUGGACACACCGGUCUUUGGUCCAGUCCAGAAGCCAAGUGCGUGGGUAAGUACAAAUCAAUGA-A- .....(((((..(.((((((-(......))))).((((((((((((....))))))))))...))((..(((((......)))))..))..))..)..)))))..............-.- ( -40.50) >DroEre_CAF1 28778 119 + 1 CAAUAUUACGUGCAACGCGG-GCUAUGUGUUGGAGGGUUCACAAUUCGCAAGUUGUGGACACACCGGAAUUUGGUCUAGCCCAGAGACCAAGUGUGUGUGUAAGUGCAAAUCCGUGGUAC ...((((((...(((((((.-....)))))))..(((((((((((......)))))).((((((....(((((((((.......)))))))))..)))))).......))))))))))). ( -39.50) >DroYak_CAF1 27635 119 + 1 CAAUAUCAGCUGCAACGCGG-GUUAUGUGUUGCAAGGUUCACAGUUCGCCAGUUGUGGACACACCGGACUUUGGUCUAGUCCAGAGACCAAGUGUGUGGGUAAGUGCAAAUCAGUGGUGC ...(((((..(((((((((.-....))))))))).(((((((.((((((.....))))))((.(((.((((((((((.......)))))))).)).)))))..)))..))))..))))). ( -40.00) >consensus CAAUAUUACCUGCAACGCGG_GUUAUAUCCUGCAGGGUUCACAAUUCGCGAGUUGUGGACACACCGGACUUUGGUCUAGUCCAGAAGCCAAGUGCGUGGGUAAGUACAAAUCAAUGA_AC .....(((((..(..(((((.((........))...((((((((((....))))))))))...))((..(((((......)))))..))..))).)..)))))................. (-26.52 = -26.72 + 0.20)

| Location | 7,732,509 – 7,732,628 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.84 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -17.47 |
| Energy contribution | -17.31 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -0.14 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7732509 119 - 23771897 UAUAAAAUCCUAUAGCACUAACUUAAAUUGUUUGUACAUUGU-ACAUUGAUGUGUACUUACCCACGCACUUGGCUUCUGGACUAGACCAAAGUCCGGUGUGUCCACAAACCGCAAAUUGU .............................((((((.....((-((((....))))))......(((((((.(((((.(((......)))))))).)))))))..)))))).......... ( -27.10) >DroSec_CAF1 28946 110 - 1 UUAAAAAUCCUAUAGCUUUAACUUCAAUUGUA---------U-UCAUUGAUUUGUACUUACCCACGCACUUGGCUUCUGGACUGGACCAAAGACCGGUGUGUCCACAACUCGCGAAUUGU ................................---------.-.....(((((((........(((((((.(((((.(((......)))))).))))))))).........))))))).. ( -18.13) >DroSim_CAF1 25074 110 - 1 UUCAAAAUCCUAUAGCUUUAACUUCGAUUGUA---------U-UCAUUGAUUUGUACUUACCCACGCACUUGGCUUCUGGACUGGACCAAAGACCGGUGUGUCCACAACUCGCGAAUUGU .........................((.....---------.-))...(((((((........(((((((.(((((.(((......)))))).))))))))).........))))))).. ( -18.23) >DroEre_CAF1 28817 115 - 1 UGCAACUUCAAUUA-----AACCAAAGUCACUCGAACCCUGUACCACGGAUUUGCACUUACACACACACUUGGUCUCUGGGCUAGACCAAAUUCCGGUGUGUCCACAACUUGCGAAUUGU .((((.........-----.....((((....((((.((........)).)))).))))((((((....(((((((.......)))))))......)))))).......))))....... ( -21.50) >DroYak_CAF1 27674 94 - 1 UGCAAAAUCUGAUA--------------------------GCACCACUGAUUUGCACUUACCCACACACUUGGUCUCUGGACUAGACCAAAGUCCGGUGUGUCCACAACUGGCGAACUGU ((((((.((.....--------------------------........)))))))).......(((((((.((.((.(((......))).)).)))))))))(((....)))........ ( -20.72) >consensus UGCAAAAUCCUAUAGC__UAACUUAAAUUGUA________GU_CCAUUGAUUUGUACUUACCCACGCACUUGGCUUCUGGACUAGACCAAAGUCCGGUGUGUCCACAACUCGCGAAUUGU ................................................(((((((........(((((((.((.((.(((......))).)).))))))))).........))))))).. (-17.47 = -17.31 + -0.16)

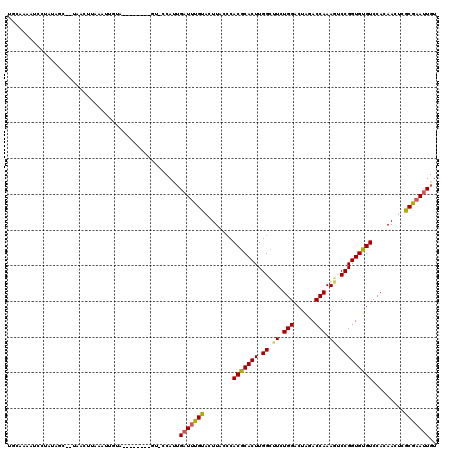
| Location | 7,732,628 – 7,732,736 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.67 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -12.06 |
| Energy contribution | -12.38 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7732628 108 - 23771897 UUGCAGCUUGCAGGAUAGAAACGUCCAUUGCGAGGGGGUGUCAAAACUGGACAACUCAAUGCGAGGGAUUCUAGAAAUAGGAUGCAAU---AGCAAAU---------ACAUAAUGUGUCU ((((..((((((((((......)))).....(((....((((.......)))).)))..))))))(.((((((....)))))).)...---.))))((---------((.....)))).. ( -30.80) >DroSec_CAF1 29056 108 - 1 UUGCAGCUGGCAGGAUAGAAACGUCCAUUGCGUGGAGGUGUCAAAACUGGACAGCUCCAUGCGAGGGAUUCUAGAAAUAUGAUGCAAU---ACCAAAU---------ACAUAAUGAGUCU (((((..........((((...((((.((((((((((.((((.......)))).)))))))))).)))))))).........))))).---.......---------............. ( -35.31) >DroSim_CAF1 25184 108 - 1 UUGCAGCUGGCAGGAUAGAAACGUCCAUUGCGUGGAGGUGUCAAAGCUGGACAGCUCCAUGCGAGGGAUUCUAGAAAUAGGAUGCAAU---ACCAAAU---------ACAUAAUGAGUCU (((((.(((......((((...((((.((((((((((.((((.......)))).)))))))))).))))))))....)))..))))).---.......---------............. ( -37.00) >DroEre_CAF1 28932 118 - 1 UGGCAGCUUGCCGGAACGAAACUUCCAUGGCUGGGUGAUUUCAAAGCGGGACAACUUAGUGCGAGCGAGUCUAUACAUAG--AUAAAUAGAUCUAUAGAUAAAGUAACUAUAAUAAGGCU .(((.((((((.((((......))))...(((((((..(..(.....)..)..)))))))))))))..(((((....(((--((......)))))))))).................))) ( -29.50) >DroYak_CAF1 27768 108 - 1 UUGCAGCUCUUCGGAUAGAAACGUCCAUGGCUGGGUGUUUCCAAAACUGGACAACUUGAUGCAAUCGAGGCUAUACAAAGGGAUAAGU---UACAAAG---------AUAUAACGAGGCU ..((..(((((.(.((((....(((((.(..((((....))))...))))))..((((((...)))))).)))).).)))))....((---((.....---------...))))...)). ( -24.10) >consensus UUGCAGCUGGCAGGAUAGAAACGUCCAUUGCGGGGAGGUGUCAAAACUGGACAACUCAAUGCGAGGGAUUCUAGAAAUAGGAUGCAAU___ACCAAAU_________ACAUAAUGAGUCU (((((.((.(((((((......))))..))).))(((.((((.......)))).)))..)))))........................................................ (-12.06 = -12.38 + 0.32)

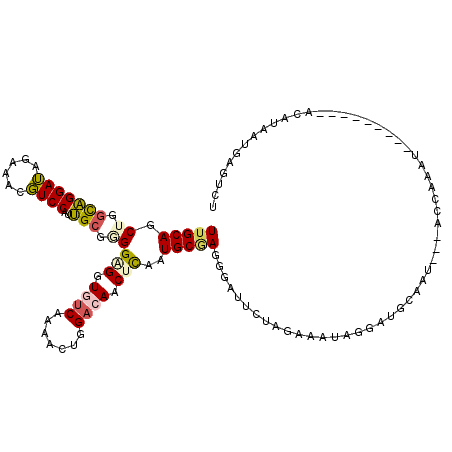
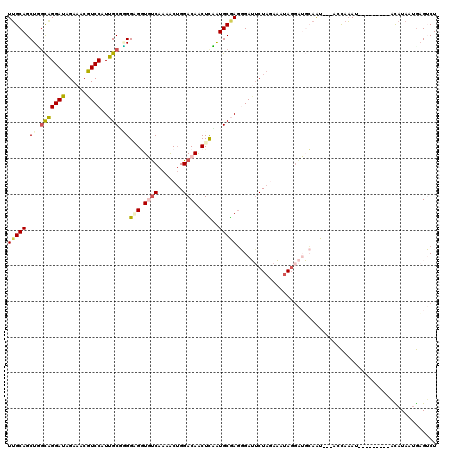
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:45 2006