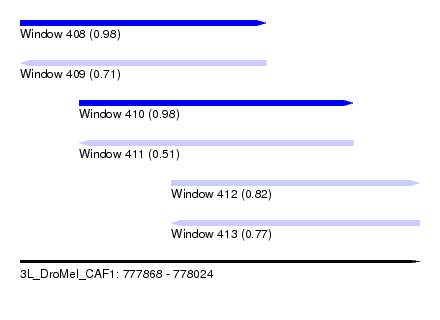
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 777,868 – 778,024 |
| Length | 156 |
| Max. P | 0.979359 |

| Location | 777,868 – 777,964 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 777868 96 + 23771897 AGAUGAUGUUUC-----CCGUU-CCCGUG----------UGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGG ......((((..-----((...-...(((----------....)))......(((.(((((((((..(((((.........)))))..))))))))).))).))..))-)).. ( -22.50) >DroVir_CAF1 155292 97 + 1 ----------------UGUGUGGCAUGUGUGUGUAUUGGUGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGCCCAAACAGG ----------------(((..((((....(((((((....))))))).....(((.(((((((((..(((((.........)))))..))))))))).)))))))...))).. ( -29.80) >DroPse_CAF1 146686 112 + 1 UGUCUGUGGUCUCUUGUCCGUGGCCUGUGUUUGUGUUUCUUAGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGG ...((((((.(....).)).(((((...(((((((((....)))))))))..(((.(((((((((..(((((.........)))))..))))))))).))).))))))-))). ( -38.40) >DroWil_CAF1 151756 100 + 1 AGUUGGAGCU-------ACAUU-UCGGUG----UGUUGGUGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGC .((((((((.-------((((.-....((----(((......))))).....))))(((((((((..(((((.........)))))..)))))))))..)))..))))-)... ( -29.30) >DroAna_CAF1 153347 88 + 1 UGGUGGUGGUG--------GU------GG----------UGGGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGG .........((--------.(------((----------(.((((((....)))(.(((((((((..(((((.........)))))..))))))))).)))).)))).-)).. ( -24.50) >DroPer_CAF1 152525 112 + 1 UGUCUGUGGUCUCUUGUCCGUGGCCUGUGUUUGUGUUUCUUAGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGG ...((((((.(....).)).(((((...(((((((((....)))))))))..(((.(((((((((..(((((.........)))))..))))))))).))).))))))-))). ( -38.40) >consensus UGUUGGUGGU_______CCGUG_CCUGUG____UGUU__UGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA_CAGG ....................................................(((.(((((((((..(((((.........)))))..))))))))).)))............ (-17.60 = -17.60 + -0.00)

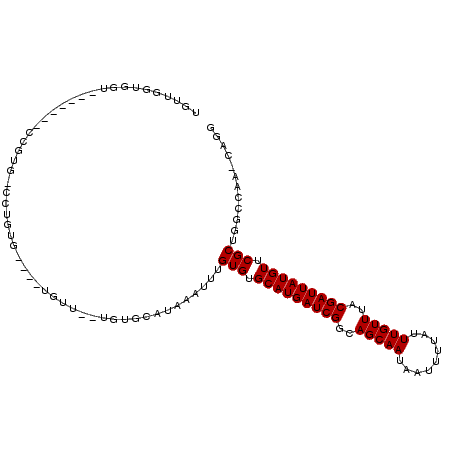
| Location | 777,868 – 777,964 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -16.25 |
| Consensus MFE | -11.80 |
| Energy contribution | -11.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 777868 96 - 23771897 CCUG-UUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCACA----------CACGGG-AACGG-----GAAACAUCAUCU ((((-(((((.(((((...((((..((......))..))))))))))))))...((((..........))))..----------..))))-....(-----....)....... ( -17.80) >DroVir_CAF1 155292 97 - 1 CCUGUUUGGGCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCACACCAAUACACACACAUGCCACACA---------------- ..(((.((((((((((...((((..((......))..)))))))))))).....((((..........))))..........)).))).........---------------- ( -16.50) >DroPse_CAF1 146686 112 - 1 CCUG-UUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCUAAGAAACACAAACACAGGCCACGGACAAGAGACCACAGACA .(((-((((((.((((......)))).........(((((..((.(((.......)))))..))))).....................))))).((........))))))... ( -18.30) >DroWil_CAF1 151756 100 - 1 GCUG-UUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCACACCAACA----CACCGA-AAUGU-------AGCUCCAACU ((((-(((((.(((((...((((..((......))..))))))))))))))...((((..........))))........----......-....)-------)))....... ( -15.30) >DroAna_CAF1 153347 88 - 1 CCUG-UUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCCCA----------CC------AC--------CACCACCACCA ...(-(((((.(((((...((((..((......))..)))))))))))))))......................----------..------..--------........... ( -11.30) >DroPer_CAF1 152525 112 - 1 CCUG-UUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCUAAGAAACACAAACACAGGCCACGGACAAGAGACCACAGACA .(((-((((((.((((......)))).........(((((..((.(((.......)))))..))))).....................))))).((........))))))... ( -18.30) >consensus CCUG_UUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCACA__AACA____CACAGG_CACGG_______ACCACCAACA ..((.((((.((((((...((((..((......))..)))))))))))))).)).(((..........))).......................................... (-11.80 = -11.80 + -0.00)

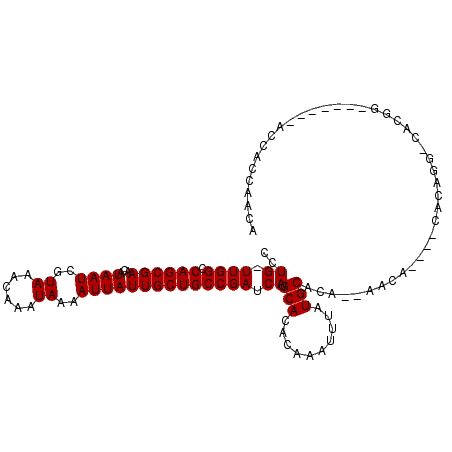
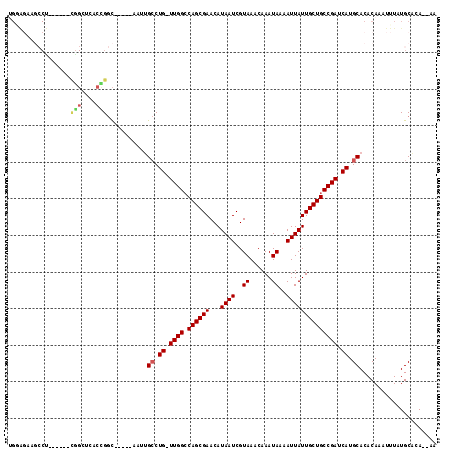
| Location | 777,891 – 777,998 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.90 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -19.85 |
| Energy contribution | -19.49 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 777891 107 + 23771897 ----UGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGGCAAUG-----GCCAGUAAGCCAGUCAGUUGGCUUCUCCA ----...(((((....)))))(((((((((..(((((.........)))))..)))))))))..((((((((.-.......))-----))))))(((((((....)))))))..... ( -36.50) >DroVir_CAF1 155311 89 + 1 UUGGUGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGCCCAAACAGGCAACA-----AGCGAU----------------------- ......(((........(((.(((((((((..(((((.........)))))..))))))))).)))((((......))))...-----.)))..----------------------- ( -25.00) >DroPse_CAF1 146721 110 + 1 UUUCUUAGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGGCUCUUGUACUGCCGGGGACCCG------AGGCGCCUCCA .((((..(((.......(((.(((((((((..(((((.........)))))..))))))))).))).((((..-..)))).......)))..))))...(------((....))).. ( -32.10) >DroWil_CAF1 151779 110 + 1 UUGGUGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGCCAAUA-----GCCAG-AUGCUGCUGACCAGGUUUGUUCC ((((((((((((......)))))))...((((((((((((((.((......)).)))))).....(((((...-.........-----)))))-.)))))))))))))......... ( -33.00) >DroAna_CAF1 153362 107 + 1 ----UGGGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGGCAAUG-----GCCAGUCAGCCGGCCAAUGGUCAGCCGCA ----..(((........(((.(((((((((..(((((.........)))))..))))))))).)))((((((.-..(((...(-----((......))).)))..)))))))))... ( -39.60) >DroPer_CAF1 152560 110 + 1 UUUCUUAGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA-CAGGCUCUUGUACUGCCGGGGACCCG------AGGCGCCUCCA .((((..(((.......(((.(((((((((..(((((.........)))))..))))))))).))).((((..-..)))).......)))..))))...(------((....))).. ( -32.10) >consensus UU__UGUGCAUAAAUUUGUGUGCAUGAUCGGCAGCAAUAAUUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAA_CAGGCAAUA_____GCCAGUAAGCCG______AGGCGCCUCCA .................(((.(((((((((..(((((.........)))))..))))))))).)))((((..................))))......................... (-19.85 = -19.49 + -0.36)

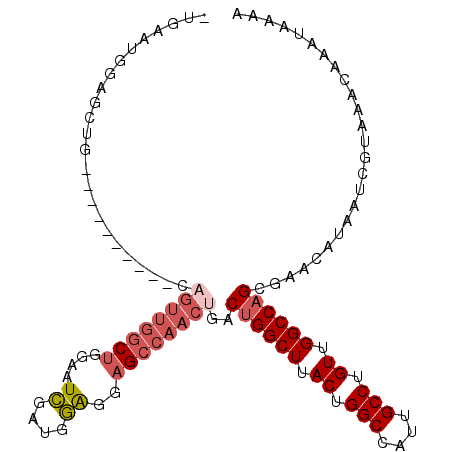
| Location | 777,891 – 777,998 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.90 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -14.50 |
| Energy contribution | -14.78 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 777891 107 - 23771897 UGGAGAAGCCAACUGACUGGCUUACUGGC-----CAUUGCCUG-UUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCACA---- (((..((((((......)))))).....)-----)).(((.((-(((((.(((((...((((..((......))..)))))))))))))).)).)))................---- ( -25.40) >DroVir_CAF1 155311 89 - 1 -----------------------AUCGCU-----UGUUGCCUGUUUGGGCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCACACCAA -----------------------...(((-----((((((.((..(.((((((((...((((..((......))..)))))))))))).).)).))).))))......))....... ( -19.50) >DroPse_CAF1 146721 110 - 1 UGGAGGCGCCU------CGGGUCCCCGGCAGUACAAGAGCCUG-UUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCUAAGAAA .((.(((.(..------..))))))((((((((...(.(((..-..)))).((((......))))................)))))))).((..(((..........)))...)).. ( -25.90) >DroWil_CAF1 151779 110 - 1 GGAACAAACCUGGUCAGCAGCAU-CUGGC-----UAUUGGCUG-UUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCACACCAA ((........(((((.((((((.-.((((-----((.......-.))))))((((......))))................)))))).)))))((((..........))))..)).. ( -30.00) >DroAna_CAF1 153362 107 - 1 UGCGGCUGACCAUUGGCCGGCUGACUGGC-----CAUUGCCUG-UUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCCCA---- .(.(((.......(((((((....)))))-----)).(((.((-(((((.(((((...((((..((......))..)))))))))))))).)).)))...........)))).---- ( -31.10) >DroPer_CAF1 152560 110 - 1 UGGAGGCGCCU------CGGGUCCCCGGCAGUACAAGAGCCUG-UUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCUAAGAAA .((.(((.(..------..))))))((((((((...(.(((..-..)))).((((......))))................)))))))).((..(((..........)))...)).. ( -25.90) >consensus UGGAGAAGCCU______CGGCUCACCGGC_____AAUUGCCUG_UUGGCCAGCGAACAUAAUCGUAAACAAAUAAAAUUAUUGCUGCCGAUCAUGCACACAAAUUUAUGCACA__AA .................(((....)))...........((.((.((((.((((((...((((..((......))..)))))))))))))).)).))..................... (-14.50 = -14.78 + 0.28)

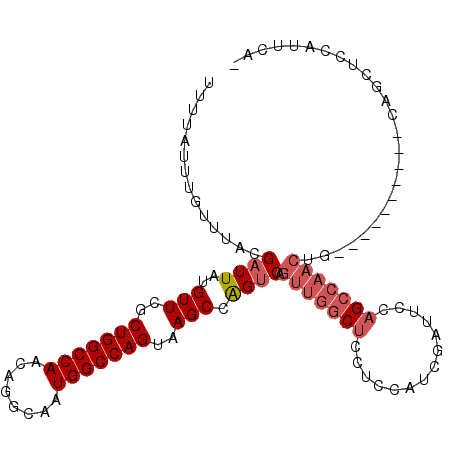
| Location | 777,927 – 778,024 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -22.81 |
| Energy contribution | -23.84 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 777927 97 + 23771897 UUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUUCUCCAUCGAUUCCAGCCAACUC----------CGGCCA---ACU- ..............((((..(((..(((((((........)))))))..))).))))((((((((((......))....)))))))).----------......---...- ( -28.80) >DroSec_CAF1 134755 100 + 1 UUUUAUUUGUUUACGAUUAUGUUCCCUGGCCAACAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCUCCAUCGAUUCCAGCCAACUG----------CAGCUCCAUUCA- ......((((....((((..(((..(((((((........)))))))..))).))))((((((((..((....))....)))))))))----------))).........- ( -27.60) >DroSim_CAF1 135245 100 + 1 UUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCUCCAUCGAUUCCAGCCAACUG----------CAGCUCCAUUCA- ......((((....((((..(((..(((((((........)))))))..))).))))((((((((..((....))....)))))))))----------))).........- ( -29.50) >DroEre_CAF1 140550 110 + 1 UUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCUCCAUCGAUUCCAGCCAACUGCGGCCAACUCCAGCUCCAUUCA- ..............((....(((.((((((((........))))))))..(((.(.(((((((((..((....))....))))))))))))).......))).....)).- ( -32.00) >DroYak_CAF1 140143 110 + 1 UUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUUCUCCAUCGAUUCCAGCCAACGCCGGCCAACUGCAACUCCAUUCA- ..............((....(((.(((((((....(((..((((......))))....(((((((((......))....)))))))))))))))...))))).....)).- ( -30.60) >DroAna_CAF1 153398 95 + 1 UUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCAAUGGCCAGUCAGCCGGCCAAUGGUCAGCCGCAGCCAACCAGGC----------------CAACUCCAUUCAU ..............((....(((.((((((((........)))))))).))).((((..((((..((....))..)))))))----------------)........)).. ( -29.40) >consensus UUUUAUUUGUUUACGAUUAUGUUCGCUGGCCAACAGGCAAUGGCCAGUAAGCCAGUCAGUUGGCUCCUCCAUCGAUUCCAGCCAACUG__________CAGCUCCAUUCA_ ..............((((..(((..(((((((........)))))))..))).)))).(((((((..............)))))))......................... (-22.81 = -23.84 + 1.03)

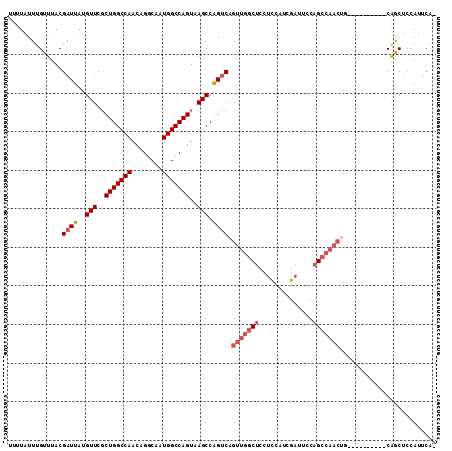
| Location | 777,927 – 778,024 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -22.51 |
| Energy contribution | -23.60 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 777927 97 - 23771897 -AGU---UGGCCG----------GAGUUGGCUGGAAUCGAUGGAGAAGCCAACUGACUGGCUUACUGGCCAUUGCCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAA -(((---.(((((----------(((((((((....((....))..))))))))..)))))).)))(((((........))))).((((......))))............ ( -36.10) >DroSec_CAF1 134755 100 - 1 -UGAAUGGAGCUG----------CAGUUGGCUGGAAUCGAUGGAGGAGCCAACUGACUGGCUUACUGGCCAUUGCCUGUUGGCCAGGGAACAUAAUCGUAAACAAAUAAAA -...((((((((.----------(((((((((....((....))..)))))))))...))))).(((((((........)))))))....))).................. ( -35.00) >DroSim_CAF1 135245 100 - 1 -UGAAUGGAGCUG----------CAGUUGGCUGGAAUCGAUGGAGGAGCCAACUGACUGGCUUACUGGCCAUUGCCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAA -...((((((((.----------(((((((((....((....))..)))))))))...))))).(((((((........)))))))....))).................. ( -34.10) >DroEre_CAF1 140550 110 - 1 -UGAAUGGAGCUGGAGUUGGCCGCAGUUGGCUGGAAUCGAUGGAGGAGCCAACUGACUGGCUUACUGGCCAUUGCCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAA -...(((..((((((((.((((.(((((((((....((....))..)))))))))...)))).)))(((....)))......)))))...))).................. ( -38.00) >DroYak_CAF1 140143 110 - 1 -UGAAUGGAGUUGCAGUUGGCCGGCGUUGGCUGGAAUCGAUGGAGAAGCCAACUGACUGGCUUACUGGCCAUUGCCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAA -....((...((((.(((((((((((((((((....((....))..)))))))....(((((....)))))......))))))))))((......)))))).))....... ( -39.20) >DroAna_CAF1 153398 95 - 1 AUGAAUGGAGUUG----------------GCCUGGUUGGCUGCGGCUGACCAUUGGCCGGCUGACUGGCCAUUGCCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAA (((((((.(((((----------------(((((((..((....))..))))..))))))))(.(((((((........))))))))...)))..))))............ ( -39.60) >consensus _UGAAUGGAGCUG__________CAGUUGGCUGGAAUCGAUGGAGGAGCCAACUGACUGGCUUACUGGCCAUUGCCUGUUGGCCAGCGAACAUAAUCGUAAACAAAUAAAA ........................((((((((....((....))..))))))))..((((((.((.(((....))).)).))))))......................... (-22.51 = -23.60 + 1.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:04 2006