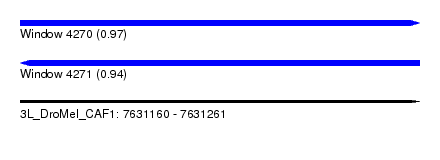
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,631,160 – 7,631,261 |
| Length | 101 |
| Max. P | 0.971559 |

| Location | 7,631,160 – 7,631,261 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -30.96 |
| Energy contribution | -30.60 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7631160 101 + 23771897 UGACUCUUCCUUGAACAGAUC---------------CUUCUUCCCGCUCCAGCUUCUGCUUUAGCAGGGCUUCGAUGCUCUCUACUUGACUUGAGGGCACCACUUCGGUGCGGGUU .....................---------------......(((((.((((((.((((....))))))))..(.(((((((..........))))))).).....)).))))).. ( -33.00) >DroPse_CAF1 23378 116 + 1 CCAUCUUCACCGACGCGGAUCCCUGCUUAGUGUCACCCUCCUCCAGCUCCAGCUUCUGCUUUAGCAGGGCUUCAAUGUUUUCCGCUUGGCCGGAGGGCACCACUUCGGUGCGGGUA ..((((.((((((.((((....))))..((((...((((((....(((..(((....)))..)))..((((.....((.....))..))))))))))...)))))))))).)))). ( -43.50) >DroSec_CAF1 16256 101 + 1 UGACUUUACCUUGAAGAAAUC---------------CUUCCUCCCGCUCCAGCUUCUGCUUCAGCAGGGCUUCAAUGCUCUCAACUUGACUUGAGGGCACCACUUCGGUGCGGGUU ............((((.....---------------))))..(((((.((((((.((((....))))))))....(((((((((......))))))))).......)).))))).. ( -36.80) >DroEre_CAF1 19805 101 + 1 UGACUUUCCCUUGAACGGAUC---------------CUUCCUCCCGCUCCAGCUUCUGCUUUAGCAGCGCUUCGAUGCUCUCUACUUGACUGGAGGGCACCACUUCGGUGCGGGUU ................(((..---------------..))).(((((.(((((..((((....)))).)))..(.(((((((((......))))))))).).....)).))))).. ( -36.10) >DroYak_CAF1 17109 101 + 1 UGACUUUGCCUUGAACGGAUC---------------CUUCCUCCCGCUCCAGCUUCUGCUUUAGCAGGGCUUCGAUACUCUCCACUUGACUGGAGGGCACCACUUCGGUGCGGGUU ................(((..---------------..))).(((((.((((((.((((....))))))))......(((((((......))))))).........)).))))).. ( -34.90) >DroPer_CAF1 25212 116 + 1 CCAUCUUCACCGACGCGGAUCCCUGCUUAGUGUCACCCUCCUCCAGCUCCAGCUUCUGCUUUAGCAGGGCUUCAAUGUUUUCCGCUUGGCCGGAGGGCACCACUUCGGUGCGGGUA ..((((.((((((.((((....))))..((((...((((((....(((..(((....)))..)))..((((.....((.....))..))))))))))...)))))))))).)))). ( -43.50) >consensus UGACUUUCCCUUGAACGGAUC_______________CUUCCUCCCGCUCCAGCUUCUGCUUUAGCAGGGCUUCAAUGCUCUCCACUUGACUGGAGGGCACCACUUCGGUGCGGGUU ..........................................(((((.((((((.((((....))))))))....(((((((((......))))))))).......)).))))).. (-30.96 = -30.60 + -0.36)

| Location | 7,631,160 – 7,631,261 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -38.15 |
| Consensus MFE | -28.34 |
| Energy contribution | -28.18 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7631160 101 - 23771897 AACCCGCACCGAAGUGGUGCCCUCAAGUCAAGUAGAGAGCAUCGAAGCCCUGCUAAAGCAGAAGCUGGAGCGGGAAGAAG---------------GAUCUGUUCAAGGAAGAGUCA ..(((((.((....((((((.(((..........))).)))))).(((.((((....))))..))))).)))))......---------------((.((.(((...))).)))). ( -33.50) >DroPse_CAF1 23378 116 - 1 UACCCGCACCGAAGUGGUGCCCUCCGGCCAAGCGGAAAACAUUGAAGCCCUGCUAAAGCAGAAGCUGGAGCUGGAGGAGGGUGACACUAAGCAGGGAUCCGCGUCGGUGAAGAUGG ((((((((((.....)))))(((((((((.(((((............))((((....))))..))).).)))))))).))))).((((..((.((...))))...))))....... ( -45.40) >DroSec_CAF1 16256 101 - 1 AACCCGCACCGAAGUGGUGCCCUCAAGUCAAGUUGAGAGCAUUGAAGCCCUGCUGAAGCAGAAGCUGGAGCGGGAGGAAG---------------GAUUUCUUCAAGGUAAAGUCA ..(((((.((.....(((((.(((((......))))).)))))..(((.((((....))))..))))).)))))..((((---------------.....))))............ ( -33.90) >DroEre_CAF1 19805 101 - 1 AACCCGCACCGAAGUGGUGCCCUCCAGUCAAGUAGAGAGCAUCGAAGCGCUGCUAAAGCAGAAGCUGGAGCGGGAGGAAG---------------GAUCCGUUCAAGGGAAAGUCA ..(((((.((....((((((.(((..........))).)))))).(((.((((....))))..))))).)))))..((..---------------..(((......)))....)). ( -34.10) >DroYak_CAF1 17109 101 - 1 AACCCGCACCGAAGUGGUGCCCUCCAGUCAAGUGGAGAGUAUCGAAGCCCUGCUAAAGCAGAAGCUGGAGCGGGAGGAAG---------------GAUCCGUUCAAGGCAAAGUCA ..(((((.((....((((((.(((((......))))).)))))).(((.((((....))))..))))).))))).(((..---------------..)))......(((...))). ( -36.60) >DroPer_CAF1 25212 116 - 1 UACCCGCACCGAAGUGGUGCCCUCCGGCCAAGCGGAAAACAUUGAAGCCCUGCUAAAGCAGAAGCUGGAGCUGGAGGAGGGUGACACUAAGCAGGGAUCCGCGUCGGUGAAGAUGG ((((((((((.....)))))(((((((((.(((((............))((((....))))..))).).)))))))).))))).((((..((.((...))))...))))....... ( -45.40) >consensus AACCCGCACCGAAGUGGUGCCCUCCAGUCAAGUGGAGAGCAUCGAAGCCCUGCUAAAGCAGAAGCUGGAGCGGGAGGAAG_______________GAUCCGUUCAAGGGAAAGUCA ..(((((.((.....(((((.(((((......))))).)))))..(((.((((....))))..))))).))))).(((...................)))................ (-28.34 = -28.18 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:21 2006