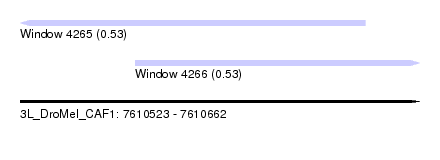
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,610,523 – 7,610,662 |
| Length | 139 |
| Max. P | 0.531206 |

| Location | 7,610,523 – 7,610,643 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.84 |
| Mean single sequence MFE | -41.03 |
| Consensus MFE | -32.08 |
| Energy contribution | -31.75 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7610523 120 - 23771897 ACUCACUUUUGUAUCUUGGGAUCCAGGCAGCUGAGCUCCUCCAGCUUGGGCAGCUCUAGACUGGAGCCAUGGCACUGCAGAUGGGCAUUCGCCUCCAUGGGCGAGUGCAUGAUGUCCAUG ..................(((((((.(((((..((((.....))))..)((.(((((.....)))))....)).))))...)))((((((((((....)))))))))).....))))... ( -45.60) >DroVir_CAF1 80979 117 - 1 ACUUACUUUUGUAUUUUUGGUUCCAAACAGCUGAGCUCCUCCAGUUUGGUCAGCUCCAGAUUGGAGGCAUGGCACUGUAGAUGGGCAUUCGCCUCCAUGGGCGAGUGCAU---GUCCAUG ...(((....)))....(((..(((.((((.((....((((((((((((......)))))))))))).....))))))...)))((((((((((....))))))))))..---..))).. ( -45.60) >DroGri_CAF1 54772 117 - 1 ACUUACUUUUGUAUUUUUGGUUCCAAACAGCUGAGCUCCUCCAGCUUGGUCAGCUCCAUAUUGGAGUCAUGGCACUGGAGAUGAGCAUUGGCCUCCAUGGGCGAGUGCAU---GUCCAUG ...(((....)))..((((....))))..((..(((((((((((....(((((((((.....)))))..)))).))))))..)))).)..))...((((((((......)---))))))) ( -40.20) >DroWil_CAF1 86970 117 - 1 ACUUACUUUUGUAUUUUUGGUUCCAAACAGCUUAGUUCCUCCAAUUUGGGUAACUCCAGACUGGAGCCAUGGCACUGCAGAUGGGCAUUUGCCUCCAUGGGUGAAUGCAU---UUCCAUA ........(((((..((((....))))..(((......(((((.((((((....)))))).)))))....)))..)))))(((((((((..(((....)))..)))))..---..)))). ( -34.00) >DroMoj_CAF1 65408 117 - 1 ACUUACUUUUGUAUUUUUGGUUCCAAACAGCUGAGCUCUUCCAGUGUGGUCAGCUCCAGGUUGGACGCAUGGCACUGCAGAUGAGCAUUCGCCUCCAUGGGCGAGUGCAU---GUCCAUG ...(((....)))..(((((........((((((.(.(.......).).))))))))))).((((((((......)))......((((((((((....))))))))))..---))))).. ( -38.50) >DroAna_CAF1 55650 117 - 1 UCUCACUUUUGUAUUUUCGGUUCCAGGCAGCUCAACUCCUCCAGCUUGGGCAGCUCUAGACUGGAACCAUGGCACUGCAAGUGGGCAUUCGCCUCCAUGGGCGAGUGCAU---GUCUAUG ..((((((..........((((((((((.((((((((.....)).)))))).))......))))))))...((...))))))))((((((((((....))))))))))..---....... ( -42.30) >consensus ACUUACUUUUGUAUUUUUGGUUCCAAACAGCUGAGCUCCUCCAGCUUGGGCAGCUCCAGACUGGAGCCAUGGCACUGCAGAUGGGCAUUCGCCUCCAUGGGCGAGUGCAU___GUCCAUG ...........(((((..(((.(((...(((...))).((((((..(((......)))..))))))...))).)))..))))).((((((((((....))))))))))............ (-32.08 = -31.75 + -0.33)

| Location | 7,610,563 – 7,610,662 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -17.73 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7610563 99 + 23771897 CUGCAGUGCCAUGGCUCCAGUCUAGAGCUGCCCAAGCUGGAGGAGCUCAGCUGCCUGGAUCCCAAGAUACAAAAGUGAGU--UCU-AUCAGCCACACUUAUU--- ..(((((.....(((....)))....)))))....(((((.((((((((.((...((.(((....))).))..)))))))--)))-.)))))..........--- ( -29.60) >DroVir_CAF1 81016 84 + 1 CUACAGUGCCAUGCCUCCAAUCUGGAGCUGACCAAACUGGAGGAGCUCAGCUGUUUGGAACCAAAAAUACAAAAGUAAGU--GCGU------------------- ..(((((......((((((.(.(((......))).).))))))......)))))(((....)))................--....------------------- ( -19.20) >DroPse_CAF1 60534 98 + 1 CUGCAGUGCCAUGGCUCCAGUCUGGAACUGCCCAAGCUGGAGGAGCUCAGCUGCCUGGACCCCAAAAUACAAAAGUAGGAAAGAA-ACUCU------UUAUCCAU ..(((((.......((((((..(((......)))..)))))).......))))).((((........(((....)))..((((..-...))------)).)))). ( -27.84) >DroGri_CAF1 54809 81 + 1 CUCCAGUGCCAUGACUCCAAUAUGGAGCUGACCAAGCUGGAGGAGCUCAGCUGUUUGGAACCAAAAAUACAAAAGUAAGU--C---------------------- (((((((.....(.((((.....))))).......)))))))..(((..(((.((((............))))))).)))--.---------------------- ( -19.00) >DroAna_CAF1 55687 93 + 1 UUGCAGUGCCAUGGUUCCAGUCUAGAGCUGCCCAAGCUGGAGGAGUUGAGCUGCCUGGAACCGAAAAUACAAAAGUGAGA--CCA-AUCCU------UUAUG--- .....(((...(((((((((..(((.(((.(((.....)).).)))....))).)))))))))....))).((((.((..--...-.))))------))...--- ( -22.80) >DroPer_CAF1 60686 98 + 1 CUGCAGUGCCAUGGCUCCAGUCUGGAACUGCCCAAGCUGGAGGAGCUCAGCUGCCUGGACCCCAAAAUACAAAAGUAGGAAAGAA-ACUCU------UUAUCCAU ..(((((.......((((((..(((......)))..)))))).......))))).((((........(((....)))..((((..-...))------)).)))). ( -27.84) >consensus CUGCAGUGCCAUGGCUCCAGUCUGGAGCUGCCCAAGCUGGAGGAGCUCAGCUGCCUGGAACCCAAAAUACAAAAGUAAGA__GCA_A__CU______UUAU____ ..(((((.......(((((((.(((......))).))))))).......))))).............(((....)))............................ (-17.73 = -17.84 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:16 2006