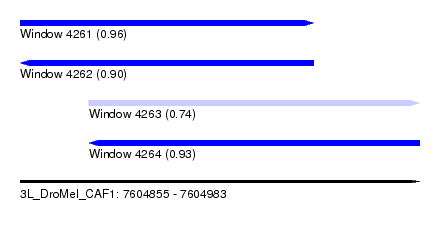
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,604,855 – 7,604,983 |
| Length | 128 |
| Max. P | 0.963217 |

| Location | 7,604,855 – 7,604,949 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -21.08 |
| Energy contribution | -21.08 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7604855 94 + 23771897 -------AACACCAAGGUGCACCAU-----UCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCAGUAUUUAUGACUUUGUGCGUGUCG -------.((((....(((((((..-----..)).)))))(((.((..((((..((......)).((((((......))))))......))))..))))))))).. ( -25.90) >DroVir_CAF1 74836 82 + 1 -------------------CACAAG-----UCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCACAAUUUAUGACUUUGUGCGUGCAC -------------------......-----.....((((((((.((..((((..((......))..(((((......))))).......))))..)))))))))). ( -24.10) >DroPse_CAF1 54297 93 + 1 GAGCGAG-ACUCCAAG-------UC-----UCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCACAAUUUAUGACUCUGUGCGUGCCG ((.((((-((.....)-------))-----))).))(((((((.....((((..((......))..(((((......))))).......))))....))))))).. ( -32.40) >DroGri_CAF1 49369 82 + 1 -------------------CACAAG-----UCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCACAAUUUAUGACUUUGUGCGUGAGU -------------------((((((-----(((((.(((........))).)))............(((((......)))))........))).)))))(....). ( -22.90) >DroAna_CAF1 50222 106 + 1 CAGCCACACCAGCAACUUGCAACAGCAACAACGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCACAAUUUAUGACUCGGUGCGUGUCG ..(((.((((......(((((...(((((....))))).)))))....((((..((......))..(((((......))))).......))))..)))).).)).. ( -27.70) >DroPer_CAF1 54424 93 + 1 GAGCGAG-ACUCCAAG-------UC-----UCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCACAAUUUAUGACUCUGUGCGUGCCG ((.((((-((.....)-------))-----))).))(((((((.....((((..((......))..(((((......))))).......))))....))))))).. ( -32.40) >consensus ________AC_CCAAG___CAC_AG_____UCGGUUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCACAAUUUAUGACUCUGUGCGUGCCG ....................................(((((((.....((((..((......))..(((((......))))).......))))....))))))).. (-21.08 = -21.08 + -0.00)



| Location | 7,604,855 – 7,604,949 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.02 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7604855 94 - 23771897 CGACACGCACAAAGUCAUAAAUACUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAACCGA-----AUGGUGCACCUUGGUGUU------- .((((((((....(((((......(((((......)))))..(((....))).))))).....))).((((.((..-----..)))))).....)))))------- ( -25.30) >DroVir_CAF1 74836 82 - 1 GUGCACGCACAAAGUCAUAAAUUGUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAACCGA-----CUUGUG------------------- (((....))).(((((........(((((......)))))....(((.(((..(((....)))..)))..))).))-----)))...------------------- ( -22.80) >DroPse_CAF1 54297 93 - 1 CGGCACGCACAGAGUCAUAAAUUGUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAACCGA-----GA-------CUUGGAGU-CUCGCUC ..(((.(((....(((((..(((((((((......)))))......))))...))))).....))).)))...(((-----((-------(.....))-))))... ( -28.70) >DroGri_CAF1 49369 82 - 1 ACUCACGCACAAAGUCAUAAAUUGUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAACCGA-----CUUGUG------------------- .......(((((.(((........(((((......)))))....(((.(((..(((....)))..)))..))).))-----))))))------------------- ( -22.30) >DroAna_CAF1 50222 106 - 1 CGACACGCACCGAGUCAUAAAUUGUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAACCGUUGUUGCUGUUGCAAGUUGCUGGUGUGGCUG .....((((((.(((.((..(((((.((((((...(((......)))..)))))).))))).(((((.(((((....)))))...))))))).))))))))).... ( -36.90) >DroPer_CAF1 54424 93 - 1 CGGCACGCACAGAGUCAUAAAUUGUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAACCGA-----GA-------CUUGGAGU-CUCGCUC ..(((.(((....(((((..(((((((((......)))))......))))...))))).....))).)))...(((-----((-------(.....))-))))... ( -28.70) >consensus CGGCACGCACAAAGUCAUAAAUUGUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAACCGA_____CU_GUG___CUUGG_GU________ ......(((....(((........(((((......))))).)))((.....((....)).....)).))).................................... (-17.02 = -17.02 + 0.00)



| Location | 7,604,877 – 7,604,983 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.18 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -19.18 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7604877 106 + 23771897 UUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCAGUAUUUAUGACUU-------UGUGCGUGUCGCACUAUCUGGAGACCUGGG--CCAUGGGAUUUCGAC---- ..(((((((.((..((((..((......)).((((((......))))))......))))..-------)))))))))...........((((((((..--...)))).))))...---- ( -26.10) >DroPse_CAF1 54318 102 + 1 UUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCACAAUUUAUGACUC-------UGUGCGUGCCGCAGUGGAGAGAGACCUCUCUCCCAG----------UUGGU ..(((((((.....((((..((......))..(((((......))))).......))))..-------..)))))))..((((((.(((((....)))))))).----------))).. ( -34.50) >DroEre_CAF1 50289 106 + 1 UUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCAGUAUUUAUGACUU-------UGUGCGUGUCUCACUAUCUGGAGACCUGGG--CCAUGGGAUUUGGAC---- ..(((((((.((..((((..((......)).((((((......))))))......))))..-------))))))))).......((..((..((....--....))..))..)).---- ( -29.60) >DroWil_CAF1 78403 95 + 1 UUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCACAAUUUAUGACUCCUCUGUGUGUGCGUGUAGCU--------------------CAUGUAGUUGGGAU---- (((((((((..........(((((....(((.(((((......)))))........)))......))))))))))))))((--------------------((......))))..---- ( -24.00) >DroAna_CAF1 50256 89 + 1 UUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCACAAUUUAUGACUC-------GGUGCGUGUCGCA-----------------------GGGAUUGAGAUUGGU ..(((((((.....((((..((......))..(((((......))))).......))))..-------..)))))))....-----------------------............... ( -20.40) >DroPer_CAF1 54445 102 + 1 UUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCACAAUUUAUGACUC-------UGUGCGUGCCGCAGUGGAGAGAGACCUCUCUCCCAG----------UUGGU ..(((((((.....((((..((......))..(((((......))))).......))))..-------..)))))))..((((((.(((((....)))))))).----------))).. ( -34.50) >consensus UUGCAUGCAAUAUUGUCACCACAUUAACGUCUUGGCCUUUUGUGGCCACAAUUUAUGACUC_______UGUGCGUGUCGCA_U_____GAGACCU_____CCAUGGGAUU__GAU____ ..(((((((.((.(((....))).))..(((.(((((......)))))........)))...........))))))).......(((((....)))))..................... (-19.18 = -18.60 + -0.58)



| Location | 7,604,877 – 7,604,983 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.18 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -16.83 |
| Energy contribution | -16.83 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7604877 106 - 23771897 ----GUCGAAAUCCCAUGG--CCCAGGUCUCCAGAUAGUGCGACACGCACA-------AAGUCAUAAAUACUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAA ----(((........((((--(....(((....))).(((((...))))).-------..)))))......(((((......))))).)))((..(((..(((....)))..))))).. ( -27.50) >DroPse_CAF1 54318 102 - 1 ACCAA----------CUGGGAGAGAGGUCUCUCUCCACUGCGGCACGCACA-------GAGUCAUAAAUUGUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAA .....----------...(((((((....)))))))......(((.(((..-------..(((((..(((((((((......)))))......))))...))))).....))).))).. ( -32.40) >DroEre_CAF1 50289 106 - 1 ----GUCCAAAUCCCAUGG--CCCAGGUCUCCAGAUAGUGAGACACGCACA-------AAGUCAUAAAUACUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAA ----..........(((((--(....((((((.....).)))))..))...-------..(((((......(((((......)))))..(((....))).))))).......))))... ( -24.10) >DroWil_CAF1 78403 95 - 1 ----AUCCCAACUACAUG--------------------AGCUACACGCACACACAGAGGAGUCAUAAAUUGUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAA ----..........((((--------------------.........(((.((((.....(((........(((((......))))).))).....))))))).........))))... ( -22.27) >DroAna_CAF1 50256 89 - 1 ACCAAUCUCAAUCCC-----------------------UGCGACACGCACC-------GAGUCAUAAAUUGUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAA ...............-----------------------((((((.((...)-------).)))....(((((.((((((...(((......)))..)))))).))))).......))). ( -20.10) >DroPer_CAF1 54445 102 - 1 ACCAA----------CUGGGAGAGAGGUCUCUCUCCACUGCGGCACGCACA-------GAGUCAUAAAUUGUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAA .....----------...(((((((....)))))))......(((.(((..-------..(((((..(((((((((......)))))......))))...))))).....))).))).. ( -32.40) >consensus ____AUC__AAUCCCAUGG_____AGGUCUC_____A_UGCGACACGCACA_______GAGUCAUAAAUUGUGGCCACAAAAGGCCAAGACGUUAAUGUGGUGACAAUAUUGCAUGCAA ..............................................(((...........(((........(((((......))))).)))((.....((....)).....)).))).. (-16.83 = -16.83 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:14 2006