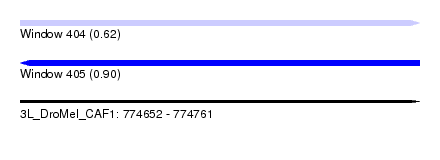
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 774,652 – 774,761 |
| Length | 109 |
| Max. P | 0.903699 |

| Location | 774,652 – 774,761 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -26.65 |
| Consensus MFE | -22.55 |
| Energy contribution | -23.05 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 774652 109 + 23771897 --GCAUCGCAUGUUGCCGGCUCAUUAGUUUGGCUUUUAUUUUUGGAAAAGCCAACGCGACGCAUUUGCCGGCCUGUCUGCCUCUGAUUUU-CCUCCGAUAUCCCCCCCUCCC --.....(((..(.((((((......(((((((((((........))))))))....)))......))))))..)..)))..........-..................... ( -25.90) >DroVir_CAF1 152057 89 + 1 CCGCAUUGCAUGUUGCCGGCUCAUUAGUUUGGCUUUUAUUUUUGGAAAAGCCAACGCGACGCAU-UGCCAGCCUGUCUGGCUCAAAAUCA---------------------- ..(((.(((..(((((..(((....)))(((((((((........))))))))).)))))))).-))).((((.....))))........---------------------- ( -26.70) >DroPse_CAF1 142149 106 + 1 CCGCAUUGCAUGUUGCCGGCUCAUUAGUUUGGCUUUUAUUUUUGGAAAAGCCAACGCGACGCAUUCGCCGGCCUGUCUGCCUAUG-UCUG-CCUCUGUCUGUCC-CUCU--- ..((((.(((..(.((((((......(((((((((((........))))))))).))((.....))))))))..)..)))..)))-)...-.............-....--- ( -28.30) >DroWil_CAF1 145204 94 + 1 CCGCAUUGCAUGUUGCCGGCUCAUUAGUUUAGCUUUUAUUUUUGGAAAAGCCAACGCGACGCAUUUGCCGGCCUGUCUGCUGUUGCUGUUGCCU------------------ ..(((..(((..(.((((((......(((..((((((........)))))).)))((...))....))))))..)..)))...)))........------------------ ( -24.80) >DroAna_CAF1 149944 101 + 1 CCGCAUUGCAUGUUGCCGGCUCAUUAGUUUGGCUUUUAUUUUUGGAAAAGCCAACGCGACGCAUUUGCCGGCCUGUCUGCCUCCGAAUUU-UUCCCCUUCUC---------- ..(((..(((....((((((......(((((((((((........))))))))....)))......)))))).))).)))..........-...........---------- ( -25.90) >DroPer_CAF1 147968 106 + 1 CCGCAUUGCAUGUUGCCGGCUCAUUAGUUUGGCUUUUAUUUUUGGAAAAGCCAACGCGACGCAUUCGCCGGCCUGUCUGCCUAUG-UCUG-CCUCUGUCUGUCC-CUCU--- ..((((.(((..(.((((((......(((((((((((........))))))))).))((.....))))))))..)..)))..)))-)...-.............-....--- ( -28.30) >consensus CCGCAUUGCAUGUUGCCGGCUCAUUAGUUUGGCUUUUAUUUUUGGAAAAGCCAACGCGACGCAUUUGCCGGCCUGUCUGCCUCUGAUUUU_CCUC_GU______________ ..(((..(((....((((((......(((((((((((........))))))))....)))......)))))).))).)))................................ (-22.55 = -23.05 + 0.50)

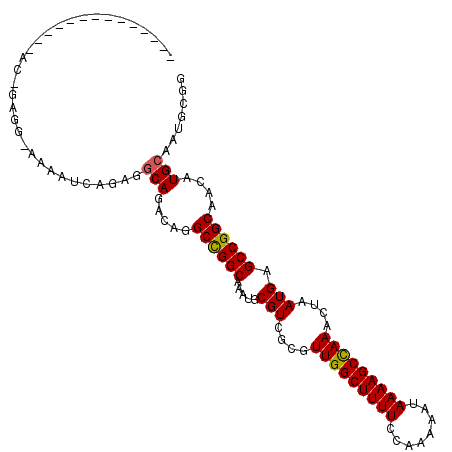
| Location | 774,652 – 774,761 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -26.63 |
| Energy contribution | -26.52 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
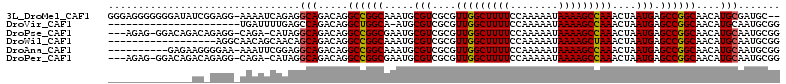
>3L_DroMel_CAF1 774652 109 - 23771897 GGGAGGGGGGGAUAUCGGAGG-AAAAUCAGAGGCAGACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCGAUGC-- ............((((.((..-....))....(((.....((((((.(((((...)))))(((((((........)))))))..........))))))....))))))).-- ( -30.40) >DroVir_CAF1 152057 89 - 1 ----------------------UGAUUUUGAGCCAGACAGGCUGGCA-AUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGG ----------------------.........(((((.....))))).-.(((((.((((((((((((........))))))))............(....))))).))))). ( -26.90) >DroPse_CAF1 142149 106 - 1 ---AGAG-GGACAGACAGAGG-CAGA-CAUAGGCAGACAGGCCGGCGAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGG ---....-............(-((..-.....(((.....((((((.....(((....(((((((((........)))))))))....))).))))))....)))..))).. ( -31.00) >DroWil_CAF1 145204 94 - 1 ------------------AGGCAACAGCAACAGCAGACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCUAAACUAAUGAGCCGGCAACAUGCAAUGCGG ------------------.(....).(((...(((.....((((((.(((((...)))))(((((((........)))))))..........))))))....)))..))).. ( -29.80) >DroAna_CAF1 149944 101 - 1 ----------GAGAAGGGGAA-AAAUUCGGAGGCAGACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGG ----------...........-.....((.(.(((.....((((((.(((((...)))))(((((((........)))))))..........))))))....)))..).)). ( -29.00) >DroPer_CAF1 147968 106 - 1 ---AGAG-GGACAGACAGAGG-CAGA-CAUAGGCAGACAGGCCGGCGAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGG ---....-............(-((..-.....(((.....((((((.....(((....(((((((((........)))))))))....))).))))))....)))..))).. ( -31.00) >consensus ______________AC_GAGG_AAAAUCAGAGGCAGACAGGCCGGCAAAUGCGUCGCGUUGGCUUUUCCAAAAAUAAAAGCCAAACUAAUGAGCCGGCAACAUGCAAUGCGG ................................(((.....((((((.....(((....(((((((((........)))))))))....))).))))))....)))....... (-26.63 = -26.52 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:57 2006