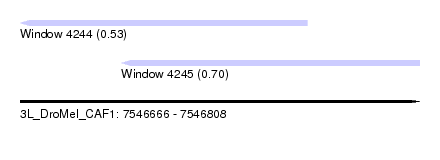
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,546,666 – 7,546,808 |
| Length | 142 |
| Max. P | 0.699782 |

| Location | 7,546,666 – 7,546,768 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 86.32 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -24.18 |
| Energy contribution | -27.18 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7546666 102 - 23771897 GUUAAGCCGAUGGGGAGGUGGGUCAUUUGUCAGCAGGAAGCUGCUGGAAUCGAGGAAAGCCACUGCCAAUGCCGCUUUUGCUGC-AAAAGCAAAAGCAAAAGG ......((.....((.(((((.(..(((((((((((....)))))))...))))...).))))).))......(((((((((..-...)))))))))....)) ( -36.60) >DroSec_CAF1 16108 102 - 1 GUUAAGCCGAUGGGGAGGUGGGUCAUUUGUCAGCAGGAAGCUGCUGGAAUCGAGGAAAGCCACUGCCAAUGCCGCUUUUGCAGC-AAAAGCAAAAGCAAAAGG ......((.....((.(((((.(..(((((((((((....)))))))...))))...).))))).))......((((((((...-....))))))))....)) ( -34.30) >DroSim_CAF1 16035 102 - 1 GUUAAGCCGAUGGGGAGGUGGGUCAUUUGUCAGCAGGAAGCUGCUGGAAUCGAGGAAAGCCACUGCCAAUGCCGCUUUUGCAGC-AAAAGCAAAAGCAAAAGG ......((.....((.(((((.(..(((((((((((....)))))))...))))...).))))).))......((((((((...-....))))))))....)) ( -34.30) >DroEre_CAF1 16239 96 - 1 GUUAAGCCGAUUGGGAGGUGGGUCAUUUGUCAGCAGGAAGCUGCUGGAAUCGAGGAAAGCCACUGCCAAUGCCGCUUUUGCUGC-------AAAAGCAAAAGG .....((..(((((..(((((.(..(((((((((((....)))))))...))))...).))))).)))))))..((((((((..-------...)))))))). ( -34.90) >DroYak_CAF1 16178 102 - 1 GUUAAGCCGAUUGGGAGGUGGGUCAUUUGUCAGCAGGAAGCUGCUGGAAUCGAGGAAAGCCACUGCCAAUGCCGCAUUUGCUGC-AUAAGCAAAAGCAAAAGG ......((.(((((..(((((.(..(((((((((((....)))))))...))))...).))))).)))))...((.((((((..-...)))))).))....)) ( -35.00) >DroAna_CAF1 8204 84 - 1 GUUAACCCG------------GUCAUUUGUCAGCAGGAAGCUGCUGGAAUUGUGGAAAGCCACUGCCGAGGAAA-------GGCAAAAAGCAAAAUGCAAAGG (((..((((------------((......(((((((....)))))))....((((....)))).)))).))...-------))).....((.....))..... ( -25.00) >consensus GUUAAGCCGAUGGGGAGGUGGGUCAUUUGUCAGCAGGAAGCUGCUGGAAUCGAGGAAAGCCACUGCCAAUGCCGCUUUUGCUGC_AAAAGCAAAAGCAAAAGG ......((.....((.(((((.((.....(((((((....))))))).......))...))))).))......((((((((........))))))))....)) (-24.18 = -27.18 + 3.00)

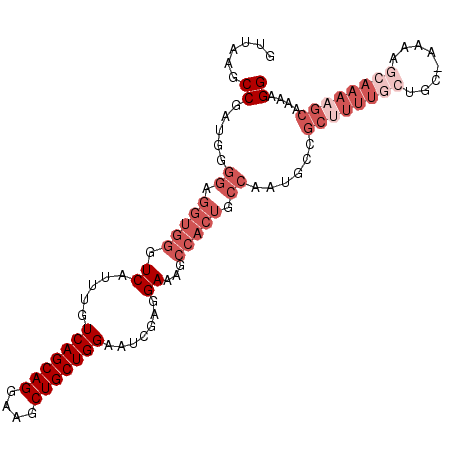

| Location | 7,546,702 – 7,546,808 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -22.47 |
| Energy contribution | -22.97 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7546702 106 - 23771897 AGAACUUUUCUUUGCCACAACAAGAAGCAAACACCAGUGGGUUAAGCCGAUGGGGAGGUGGGUCAUUUGUCAGCAGGAAGCUGCUGGAAUCGAGGAAAGCCACUGC ....((((((((.......(((((..((..((.((.((.((.....)).)).))...))..))..)))))((((((....)))))).....))))))))....... ( -28.90) >DroSec_CAF1 16144 106 - 1 AGAACUUUCCUUGGCCACAGCAAGAAGCAAACACCAGUGGGUUAAGCCGAUGGGGAGGUGGGUCAUUUGUCAGCAGGAAGCUGCUGGAAUCGAGGAAAGCCACUGC ....((((((((((((((.((.....)).....((.((.((.....)).)).))...))))........(((((((....)))))))..))))))))))....... ( -37.30) >DroSim_CAF1 16071 106 - 1 AGAACUUUCCUUGGCCACAGCAAGAAGCAAACACCAGUGGGUUAAGCCGAUGGGGAGGUGGGUCAUUUGUCAGCAGGAAGCUGCUGGAAUCGAGGAAAGCCACUGC ....((((((((((((((.((.....)).....((.((.((.....)).)).))...))))........(((((((....)))))))..))))))))))....... ( -37.30) >DroEre_CAF1 16269 89 - 1 ---------------GGCA--AAGAACCAAACACCAGUGGGUUAAGCCGAUUGGGAGGUGGGUCAUUUGUCAGCAGGAAGCUGCUGGAAUCGAGGAAAGCCACUGC ---------------(((.--..((.(((....(((((.((.....)).)))))....))).)).....(((((((....)))))))...........)))..... ( -32.90) >DroYak_CAF1 16214 98 - 1 --------CUGUCGCGACAACAAGAACCAAACACCAGUGGGUUAAGCCGAUUGGGAGGUGGGUCAUUUGUCAGCAGGAAGCUGCUGGAAUCGAGGAAAGCCACUGC --------..((.(((((((...((.(((....(((((.((.....)).)))))....))).))..)))))(((((....))))).............)).))... ( -31.30) >DroAna_CAF1 8234 94 - 1 AGAGCUCUGCGAGUUAUUGGCAAGAACCAAUUGUCAGUGGGUUAACCCG------------GUCAUUUGUCAGCAGGAAGCUGCUGGAAUUGUGGAAAGCCACUGC ..(((((...)))))((((((((.......))))))))(((....))).------------........(((((((....)))))))....((((....))))... ( -29.40) >consensus AGAACUUUCCUUGGCCACAGCAAGAACCAAACACCAGUGGGUUAAGCCGAUGGGGAGGUGGGUCAUUUGUCAGCAGGAAGCUGCUGGAAUCGAGGAAAGCCACUGC ..................................((((((.((..(((........)))...((.....(((((((....))))))).......)))).)))))). (-22.47 = -22.97 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:56 2006