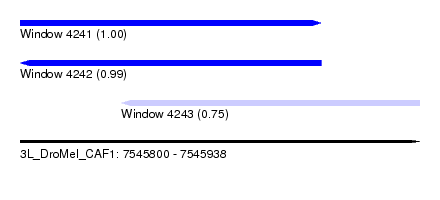
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,545,800 – 7,545,938 |
| Length | 138 |
| Max. P | 0.995054 |

| Location | 7,545,800 – 7,545,904 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -21.74 |
| Consensus MFE | -20.86 |
| Energy contribution | -20.54 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995054 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7545800 104 + 23771897 GUAUUUUCUACUAUUUCCAAACUGACCGAUAUUUGAUCUCUAUAAACCCGGUUCCGCUCCCACAUUUCGGAUUGAUUCCCAUCCAAAUCGGAGCGCUUGUCAGU ....................((((((.(((.....)))................((((((...((((.((((.(....).)))))))).))))))...)))))) ( -20.70) >DroSec_CAF1 15258 104 + 1 AUAUUUCCGACUAUUUCCAAACUGACCGAUAUUUGAUCUCCAUAAACACGGUUCCGCUCCCACAUUUCGGAUUGAUUCCCAUCCAAAUCGGAGCGCCUGUCAGU ....................((((((.(((.....)))...........((...((((((...((((.((((.(....).)))))))).)))))))).)))))) ( -21.60) >DroSim_CAF1 15176 104 + 1 AUAUUUUUGACUAUUUCCAAACUGACCGAUAUUUGAUCUCCAUAAACCCGGUUCCGCUCCCACAUUUCGGAUUGAUUCCCAUCCAAAUCGGAGCGCUUGUCAGU ......(((((............(((((.....((.....))......))))).((((((...((((.((((.(....).)))))))).))))))...))))). ( -21.80) >DroEre_CAF1 15407 104 + 1 CCAUAUCCGACUAUUUCCGAACUGACCGAUAUUUGAUCUCCAUACACCCGGUUCCGCUCCCACAUUUCGGAUUGAUUCCCAUCCAAAUCGGAGCGCUUGUCAGU ....................((((((........((...((........)).))((((((...((((.((((.(....).)))))))).))))))...)))))) ( -21.10) >DroYak_CAF1 15314 104 + 1 UUAUUUUCGACUAUUUCCCAGCUGACCGAUAUUUGAUGACCACAAACCCGGUUCCGCUCCCACAUUUCGGAUUGAUUCCCAUCCAAAUCGGAGCGCUUGUCAGU ....................((((((...........((((........)))).((((((...((((.((((.(....).)))))))).))))))...)))))) ( -23.50) >consensus AUAUUUUCGACUAUUUCCAAACUGACCGAUAUUUGAUCUCCAUAAACCCGGUUCCGCUCCCACAUUUCGGAUUGAUUCCCAUCCAAAUCGGAGCGCUUGUCAGU ....................((((((........((...((........)).))((((((...((((.((((.(....).)))))))).))))))...)))))) (-20.86 = -20.54 + -0.32)

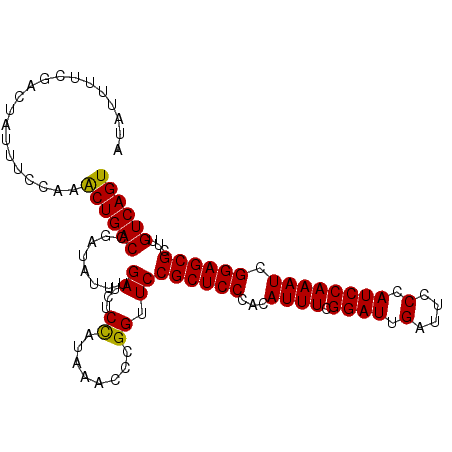
| Location | 7,545,800 – 7,545,904 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -25.96 |
| Energy contribution | -26.36 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7545800 104 - 23771897 ACUGACAAGCGCUCCGAUUUGGAUGGGAAUCAAUCCGAAAUGUGGGAGCGGAACCGGGUUUAUAGAGAUCAAAUAUCGGUCAGUUUGGAAAUAGUAGAAAAUAC ((((((...((((((.((((((((........))))))...)).))))))......(((((....)))))........)))))).................... ( -27.80) >DroSec_CAF1 15258 104 - 1 ACUGACAGGCGCUCCGAUUUGGAUGGGAAUCAAUCCGAAAUGUGGGAGCGGAACCGUGUUUAUGGAGAUCAAAUAUCGGUCAGUUUGGAAAUAGUCGGAAAUAU .(((((...((((((.((((((((........))))))...)).))))))...((((....)))).((((.......))))............)))))...... ( -30.30) >DroSim_CAF1 15176 104 - 1 ACUGACAAGCGCUCCGAUUUGGAUGGGAAUCAAUCCGAAAUGUGGGAGCGGAACCGGGUUUAUGGAGAUCAAAUAUCGGUCAGUUUGGAAAUAGUCAAAAAUAU ..((((...((((((.((((((((........))))))...)).))))))...(((..((.((.((.........)).)).))..))).....))))....... ( -28.80) >DroEre_CAF1 15407 104 - 1 ACUGACAAGCGCUCCGAUUUGGAUGGGAAUCAAUCCGAAAUGUGGGAGCGGAACCGGGUGUAUGGAGAUCAAAUAUCGGUCAGUUCGGAAAUAGUCGGAUAUGG .(((((...((((((.((((((((........))))))...)).))))))...(((..(..((.((.........)).))..)..))).....)))))...... ( -33.10) >DroYak_CAF1 15314 104 - 1 ACUGACAAGCGCUCCGAUUUGGAUGGGAAUCAAUCCGAAAUGUGGGAGCGGAACCGGGUUUGUGGUCAUCAAAUAUCGGUCAGCUGGGAAAUAGUCGAAAAUAA ..((((...((((((.((((((((........))))))...)).))))))...((.(((...((..(..........)..))))).)).....))))....... ( -29.80) >consensus ACUGACAAGCGCUCCGAUUUGGAUGGGAAUCAAUCCGAAAUGUGGGAGCGGAACCGGGUUUAUGGAGAUCAAAUAUCGGUCAGUUUGGAAAUAGUCGAAAAUAU ((((((...((((((.((((((((........))))))...)).))))))...(((......))).............)))))).................... (-25.96 = -26.36 + 0.40)

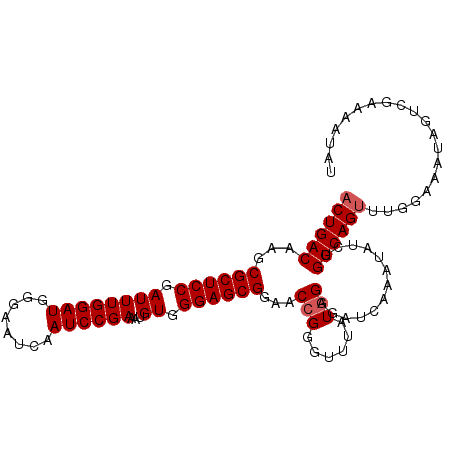
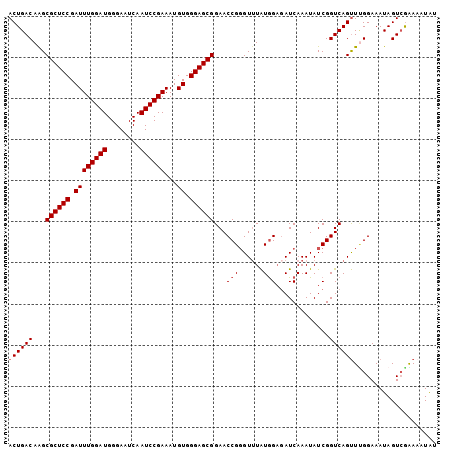
| Location | 7,545,835 – 7,545,938 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -20.00 |
| Energy contribution | -21.78 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
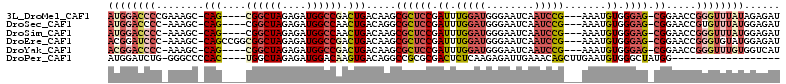
>3L_DroMel_CAF1 7545835 103 - 23771897 AUGGACCCCGAAAGC-CAG----CGGCUAGAGAUGGCCGACUGACAAGCGCUCCGAUUUGGAUGGGAAUCAAUCCG---AAAUGUGGGAG-CGGAACCGGGUUUAUAGAGAU ((((((((.(...(.-(((----((((((....)))))).))).)...((((((.((((((((........)))))---)...)).))))-))...).))))))))...... ( -41.10) >DroSec_CAF1 15293 102 - 1 AUGGACCCC-AAAGC-CAG----CGGCUAGAGAUGGCCAACUGACAGGCGCUCCGAUUUGGAUGGGAAUCAAUCCG---AAAUGUGGGAG-CGGAACCGUGUUUAUGGAGAU .......((-(((((-(((----.(((((....)))))..)))...((((((((.((((((((........)))))---)...)).))))-))...))..)))).))).... ( -31.80) >DroSim_CAF1 15211 102 - 1 AUGGACCCC-AAAGC-CAG----CGGCUAGAGAUGGCCAACUGACAAGCGCUCCGAUUUGGAUGGGAAUCAAUCCG---AAAUGUGGGAG-CGGAACCGGGUUUAUGGAGAU ((((((((.-...(.-(((----.(((((....)))))..))).)...((((((.((((((((........)))))---)...)).))))-)).....))))))))...... ( -36.70) >DroEre_CAF1 15442 106 - 1 ACGGAUCCC-AAAGC-CAGCCGGCGGCUAGAGAUGGCCGACUGACAAGCGCUCCGAUUUGGAUGGGAAUCAAUCCG---AAAUGUGGGAG-CGGAACCGGGUGUAUGGAGAU ....(((((-(....-((.((((((((((....)))))).........((((((.((((((((........)))))---)...)).))))-))...)))).))..))).))) ( -40.60) >DroYak_CAF1 15349 102 - 1 ACGGACCCC-AAAGC-CAG----CGGCUAGAGAUGGCCGACUGACAAGCGCUCCGAUUUGGAUGGGAAUCAAUCCG---AAAUGUGGGAG-CGGAACCGGGUUUGUGGUCAU ...((((.(-(((.(-(((----((((((....)))))).))......((((((.((((((((........)))))---)...)).))))-)).....)).)))).)))).. ( -43.20) >DroPer_CAF1 9620 89 - 1 AUGGAUCUG-GGGCCCCAC----UGGCUAGAGAUGGACAAGUGACAGGCCGCGCGACUCUCAAGAGAUUGAAACAGCUUGAAUGUGGGCUAUGG------------------ .........-.((((((((----(..(((....)))...)))).(((((.(..((((((....))).)))...).))))).....)))))....------------------ ( -24.40) >consensus AUGGACCCC_AAAGC_CAG____CGGCUAGAGAUGGCCAACUGACAAGCGCUCCGAUUUGGAUGGGAAUCAAUCCG___AAAUGUGGGAG_CGGAACCGGGUUUAUGGAGAU ((((((((........(((....((((((....)))))).))).....((((((.((.(((((........))))).......)).)))).)).....))))))))...... (-20.00 = -21.78 + 1.78)

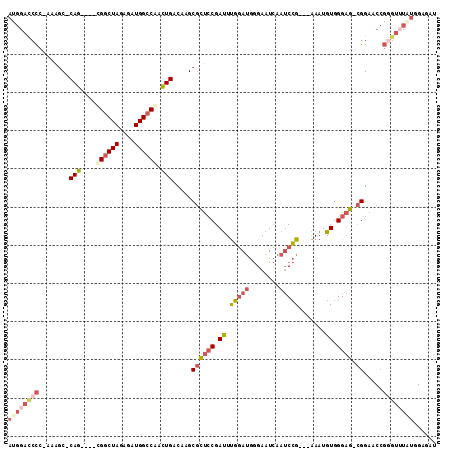
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:55 2006