
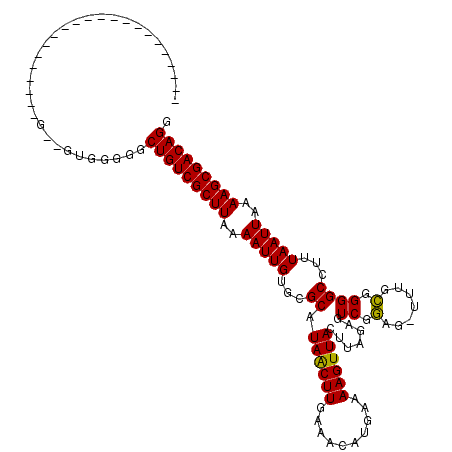
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,544,080 – 7,544,174 |
| Length | 94 |
| Max. P | 0.937083 |

| Location | 7,544,080 – 7,544,174 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -19.68 |
| Energy contribution | -19.18 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7544080 94 + 23771897 -------------------------GUGGGGGCUGUCGCUUAAAAUUGUGCGCAUAACUUGGAACAUGAAAAGUUACUUAGAGUCGGAG-UUUGCGGGGCCUUUAAUUAAAAGCGACAGG -------------------------.......(((((((((..((((((.((((.(((((.((...(((........)))...)).)))-)))))).).....)))))..))))))))). ( -27.10) >DroPse_CAF1 7376 120 + 1 AGCAGCAGAGCGGCAGAGCAGUGUUGGUGGUGCUGUCGCUUAAAAUUGUACGCAUAGCUUGAAACAUGAAAAGUUACUUAGAGUCUGAGCUUUGCGGGGCCUUUAAUUAAAAGCGACAGG ..(((((..((......))..)))))......(((((((((..(((((..((((.(((((((....(((........)))...)).))))).)))).......)))))..))))))))). ( -34.70) >DroGri_CAF1 7873 91 + 1 ----------------------UGUGCUGCUGCUGUCGCUUAAAAUUGUGCGCAUAACUUAAAACAUGAAAAGUUACUUAGAGUCGC-------GGGGGCCUUUAAUUAAAAGCGACAGG ----------------------..........(((((((((..(((((...((.((((((..........))))))((..(.....)-------..))))...)))))..))))))))). ( -20.00) >DroYak_CAF1 13503 94 + 1 -------------------------GUGGGGGCUGUCGCUUAAAAUUGUGCGCAUAACUUGGAACAUGAAAAGUUACUUAGAGUCGGAG-UUUGCGGGGCCUUUAAUUAAAAGCGACAGG -------------------------.......(((((((((..((((((.((((.(((((.((...(((........)))...)).)))-)))))).).....)))))..))))))))). ( -27.10) >DroAna_CAF1 5314 96 + 1 ---------------------UG--UUGGGGGCUGUCGCUUAAAAUUGUGCGCAUAACUUGGAACAUGAAAAGUUACUUAGAGUCGGAG-UUAGCGGGGCCUUUAAUUAAAAGCGACAGG ---------------------..--.......(((((((((..(((((.((.(.((((((.((...(((........)))...)).)))-)))...).))...)))))..))))))))). ( -28.60) >DroPer_CAF1 7423 120 + 1 AGCAGCAGAGCGGCAGAGCAGUGUUGGUGGUGCUGUCGCUUAAAAUUGUACGCAUAGCUUGAAACAUGAAAAGUUACUUAGAGUCUGAGCUUUGCGGGGCCUUUAAUUAAAAGCGACAGG ..(((((..((......))..)))))......(((((((((..(((((..((((.(((((((....(((........)))...)).))))).)))).......)))))..))))))))). ( -34.70) >consensus ______________________G__GUGGGGGCUGUCGCUUAAAAUUGUGCGCAUAACUUGAAACAUGAAAAGUUACUUAGAGUCGGAG_UUUGCGGGGCCUUUAAUUAAAAGCGACAGG ................................(((((((((..(((((...((.((((((..........)))))).......((.(.......).))))...)))))..))))))))). (-19.68 = -19.18 + -0.50)


| Location | 7,544,080 – 7,544,174 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -16.70 |
| Energy contribution | -17.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7544080 94 - 23771897 CCUGUCGCUUUUAAUUAAAGGCCCCGCAAA-CUCCGACUCUAAGUAACUUUUCAUGUUCCAAGUUAUGCGCACAAUUUUAAGCGACAGCCCCCAC------------------------- .(((((((((..((((....((...(((((-((..(((...(((.....)))...)))...)))).)))))..))))..))))))))).......------------------------- ( -20.30) >DroPse_CAF1 7376 120 - 1 CCUGUCGCUUUUAAUUAAAGGCCCCGCAAAGCUCAGACUCUAAGUAACUUUUCAUGUUUCAAGCUAUGCGUACAAUUUUAAGCGACAGCACCACCAACACUGCUCUGCCGCUCUGCUGCU .(((((((((..((((...(....)(((.((((.((((...(((.....)))...))))..)))).)))....))))..)))))))))......((.((..((......))..)).)).. ( -26.30) >DroGri_CAF1 7873 91 - 1 CCUGUCGCUUUUAAUUAAAGGCCCCC-------GCGACUCUAAGUAACUUUUCAUGUUUUAAGUUAUGCGCACAAUUUUAAGCGACAGCAGCAGCACA---------------------- .(((((((((..((((...((...))-------(((.......(((((((..........))))))).)))..))))..)))))))))..........---------------------- ( -22.60) >DroYak_CAF1 13503 94 - 1 CCUGUCGCUUUUAAUUAAAGGCCCCGCAAA-CUCCGACUCUAAGUAACUUUUCAUGUUCCAAGUUAUGCGCACAAUUUUAAGCGACAGCCCCCAC------------------------- .(((((((((..((((....((...(((((-((..(((...(((.....)))...)))...)))).)))))..))))..))))))))).......------------------------- ( -20.30) >DroAna_CAF1 5314 96 - 1 CCUGUCGCUUUUAAUUAAAGGCCCCGCUAA-CUCCGACUCUAAGUAACUUUUCAUGUUCCAAGUUAUGCGCACAAUUUUAAGCGACAGCCCCCAA--CA--------------------- .(((((((((..((((....((...(((((-((..(((...(((.....)))...)))...))))).))))..))))..))))))))).......--..--------------------- ( -20.70) >DroPer_CAF1 7423 120 - 1 CCUGUCGCUUUUAAUUAAAGGCCCCGCAAAGCUCAGACUCUAAGUAACUUUUCAUGUUUCAAGCUAUGCGUACAAUUUUAAGCGACAGCACCACCAACACUGCUCUGCCGCUCUGCUGCU .(((((((((..((((...(....)(((.((((.((((...(((.....)))...))))..)))).)))....))))..)))))))))......((.((..((......))..)).)).. ( -26.30) >consensus CCUGUCGCUUUUAAUUAAAGGCCCCGCAAA_CUCCGACUCUAAGUAACUUUUCAUGUUCCAAGUUAUGCGCACAAUUUUAAGCGACAGCACCAAC__C______________________ .(((((((((..((((...(....)..................(((((((..........)))))))......))))..)))))))))................................ (-16.70 = -17.03 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:52 2006