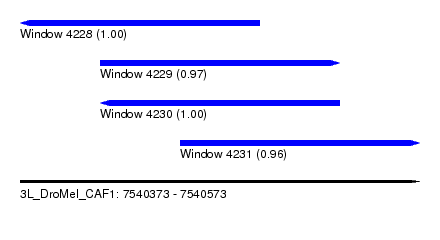
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,540,373 – 7,540,573 |
| Length | 200 |
| Max. P | 0.999958 |

| Location | 7,540,373 – 7,540,493 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -44.88 |
| Consensus MFE | -41.20 |
| Energy contribution | -41.24 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7540373 120 - 23771897 GGGACAGGCGCACUGAAAGCCACCAGGCUCGUUCUGGCAAAAGUGGGAGCAAUUCCCAUUGGCCAGAGAGCACUCAUCAAUGUCCAAGCAUGUAUGCGCUGAUGGGGUUUCGGAUGAGGU ((....(((.........))).))..((((..((((((...((((((((...)))))))).)))))))))).((((((....(((.(((........)))....))).....)))))).. ( -45.00) >DroSec_CAF1 9582 120 - 1 GGGACAGGCGCACUGAAAGCCACCUGGUUCAUUCUGACAAAAGUGGGAGCAAUUCCCAUUGGACAGAGAGCACUCAUCAAUGUCCAAGCAUGUAGGCGCUGAUGGGGCUUCGGAUGAGGU ......(((.........)))((((.((((..((((.(...((((((((...)))))))).).))))((((.(((((((.((((..........)))).))))))))))).)))).)))) ( -42.10) >DroSim_CAF1 9491 120 - 1 GGGACAGGCGCACUGAAAGCCACCUGGCUCAUUCUGACAAAAGUGGGAGCAAUUCCCAUUGGACAGAGAGCACUCAUCAAUGUCCAAGCAUGUAGGCGCUGAUGGGGCUUCGGAUGAGGU ....((((.((.......))..))))((((..((((.(...((((((((...)))))))).).)))))))).((((((...((((.(((........)))....))))....)))))).. ( -42.40) >DroEre_CAF1 9699 120 - 1 AGGACAGGCGCACAGGAAGCCACCUGGCUCAUUCUGGCAAAAGUGGGAGCAAUUCCCAUUGGCCAGGGAGCACUCAUCAAUGUCCAAGCAUGCAGGCGGAGAUGGGGCAUCGGAUGAGGU ....((((.((.......))..))))((((...(((((...((((((((...)))))))).))))).)))).((((((.((((((..((......)).......))))))..)))))).. ( -48.00) >DroYak_CAF1 9587 120 - 1 AGGACAGGCGCACUGGAAGCCACCUGGCUCAUUCUGGCAAAAGUGGGAGCAAUUCCCAUUGGCCAGCGAGCACUCAUCAAUGUCCAAGCAUGUAGGCGGAGAUGGGACAUCGGGUGAGGU ....((((.((.......))..))))((((...(((((...((((((((...)))))))).))))).)))).((((((.((((((..((......)).......))))))..)))))).. ( -46.90) >consensus GGGACAGGCGCACUGAAAGCCACCUGGCUCAUUCUGGCAAAAGUGGGAGCAAUUCCCAUUGGCCAGAGAGCACUCAUCAAUGUCCAAGCAUGUAGGCGCUGAUGGGGCUUCGGAUGAGGU ....((((.((.......))..))))((((..((((((...((((((((...)))))))).)))))))))).((((((...((((..((......)).......))))....)))))).. (-41.20 = -41.24 + 0.04)

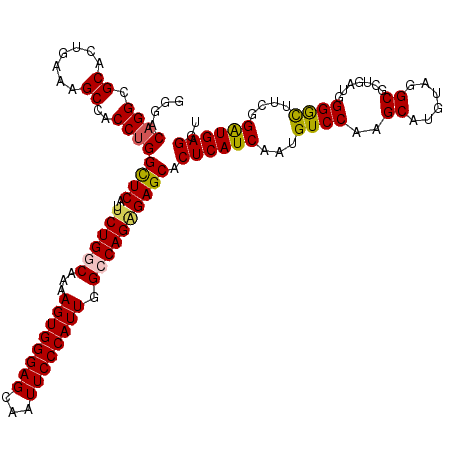
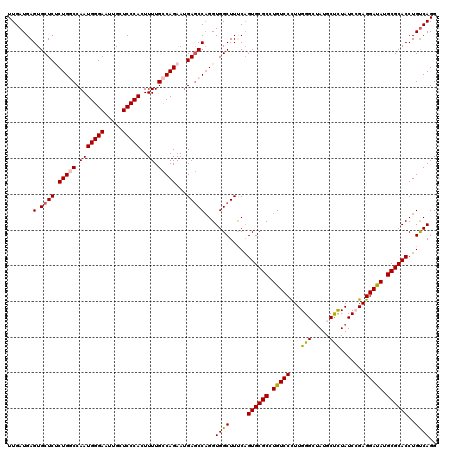
| Location | 7,540,413 – 7,540,533 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -45.42 |
| Consensus MFE | -37.98 |
| Energy contribution | -37.78 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7540413 120 + 23771897 UUGAUGAGUGCUCUCUGGCCAAUGGGAAUUGCUCCCACUUUUGCCAGAACGAGCCUGGUGGCUUUCAGUGCGCCUGUCCCUUGGGCUAUGCCCUAUCCGAGGAUAUGCGCACCUGUCAGG ...((.((.((((((((((.(((((((.....)))))..)).))))))..)))))).))..((..((((((((.((((((..((((...)))).....).))))).))))).)))..)). ( -48.10) >DroSec_CAF1 9622 120 + 1 UUGAUGAGUGCUCUCUGUCCAAUGGGAAUUGCUCCCACUUUUGUCAGAAUGAACCAGGUGGCUUUCAGUGCGCCUGUCCCUUGGGCCAUGCUCUAUCCGAGGAUAUGCGCACCUGUCAGG (((((..((((.....((((((.((((...((...(((((...((.....))...)))))((.......))))...))))))))))...((..((((....)))).))))))..))))). ( -38.90) >DroSim_CAF1 9531 120 + 1 UUGAUGAGUGCUCUCUGUCCAAUGGGAAUUGCUCCCACUUUUGUCAGAAUGAGCCAGGUGGCUUUCAGUGCGCCUGUCCCUUGGGCUAUGCUCUAUCCGAGGAUAUGCGCACCUGUCAGG (((((..((((.....((((((.((((...((((...((......))...))))((((((((.....)).))))))))))))))))...((..((((....)))).))))))..))))). ( -42.50) >DroEre_CAF1 9739 120 + 1 UUGAUGAGUGCUCCCUGGCCAAUGGGAAUUGCUCCCACUUUUGCCAGAAUGAGCCAGGUGGCUUCCUGUGCGCCUGUCCUUUGGGAUAUGCCUUAUCAGAGGACAUGCGCACCUGCCAGG .......(.((((.(((((.(((((((.....)))))..)).)))))...)))))...((((.....((((((.((((((((((...........)))))))))).))))))..)))).. ( -50.10) >DroYak_CAF1 9627 120 + 1 UUGAUGAGUGCUCGCUGGCCAAUGGGAAUUGCUCCCACUUUUGCCAGAAUGAGCCAGGUGGCUUCCAGUGCGCCUGUCCUUUGGGAUAUGCUUUAUCAGAGGAUAUGCGCACCUGUCAGG (((((..(.((((.(((((.(((((((.....)))))..)).)))))...))))).((......)).((((((.((((((((((...........)))))))))).))))))..))))). ( -47.50) >consensus UUGAUGAGUGCUCUCUGGCCAAUGGGAAUUGCUCCCACUUUUGCCAGAAUGAGCCAGGUGGCUUUCAGUGCGCCUGUCCCUUGGGCUAUGCUCUAUCCGAGGAUAUGCGCACCUGUCAGG .......(.((((.(((((.(((((((.....)))))..)).)))))...)))))...((((.....((((((.(((((...(((.....))).......))))).))))))..)))).. (-37.98 = -37.78 + -0.20)

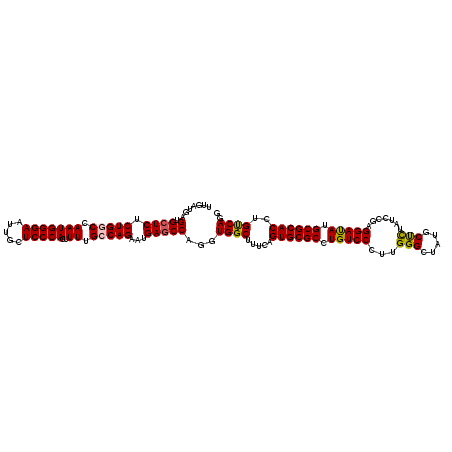
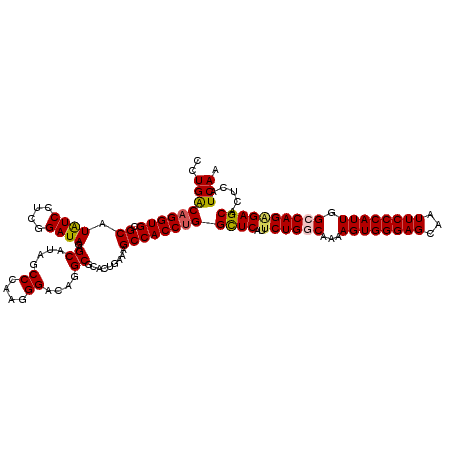
| Location | 7,540,413 – 7,540,533 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -45.48 |
| Consensus MFE | -41.58 |
| Energy contribution | -41.78 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.87 |
| SVM RNA-class probability | 0.999958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7540413 120 - 23771897 CCUGACAGGUGCGCAUAUCCUCGGAUAGGGCAUAGCCCAAGGGACAGGCGCACUGAAAGCCACCAGGCUCGUUCUGGCAAAAGUGGGAGCAAUUCCCAUUGGCCAGAGAGCACUCAUCAA ..(((..(((((((.((((....))))((((...)))).........)))))))............((((..((((((...((((((((...)))))))).)))))))))).....))). ( -48.60) >DroSec_CAF1 9622 120 - 1 CCUGACAGGUGCGCAUAUCCUCGGAUAGAGCAUGGCCCAAGGGACAGGCGCACUGAAAGCCACCUGGUUCAUUCUGACAAAAGUGGGAGCAAUUCCCAUUGGACAGAGAGCACUCAUCAA ..(((..(((((((.((((....))))..)).((..(((.((....(((.........))).)))))..))(((((.(...((((((((...)))))))).).))))).)))))..))). ( -39.80) >DroSim_CAF1 9531 120 - 1 CCUGACAGGUGCGCAUAUCCUCGGAUAGAGCAUAGCCCAAGGGACAGGCGCACUGAAAGCCACCUGGCUCAUUCUGACAAAAGUGGGAGCAAUUCCCAUUGGACAGAGAGCACUCAUCAA ..(((((((((.((.((((....))))(.((....((....))....)).).......))))))))((((..((((.(...((((((((...)))))))).).)))))))).....))). ( -41.40) >DroEre_CAF1 9739 120 - 1 CCUGGCAGGUGCGCAUGUCCUCUGAUAAGGCAUAUCCCAAAGGACAGGCGCACAGGAAGCCACCUGGCUCAUUCUGGCAAAAGUGGGAGCAAUUCCCAUUGGCCAGGGAGCACUCAUCAA ..((((..((((((.((((((..((((.....))))....)))))).)))))).....))))....((((...(((((...((((((((...)))))))).))))).))))......... ( -51.80) >DroYak_CAF1 9627 120 - 1 CCUGACAGGUGCGCAUAUCCUCUGAUAAAGCAUAUCCCAAAGGACAGGCGCACUGGAAGCCACCUGGCUCAUUCUGGCAAAAGUGGGAGCAAUUCCCAUUGGCCAGCGAGCACUCAUCAA ..(((((((((.((...(((..((.....((...(((....)))...)).))..))).))))))))((((...(((((...((((((((...)))))))).))))).)))).....))). ( -45.80) >consensus CCUGACAGGUGCGCAUAUCCUCGGAUAGAGCAUAGCCCAAGGGACAGGCGCACUGAAAGCCACCUGGCUCAUUCUGGCAAAAGUGGGAGCAAUUCCCAUUGGCCAGAGAGCACUCAUCAA ..(((((((((.((.((((....))))..((....((....))....)).........))))))))((((..((((((...((((((((...)))))))).)))))))))).....))). (-41.58 = -41.78 + 0.20)

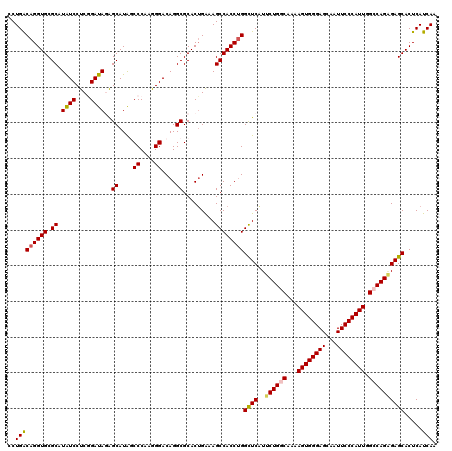
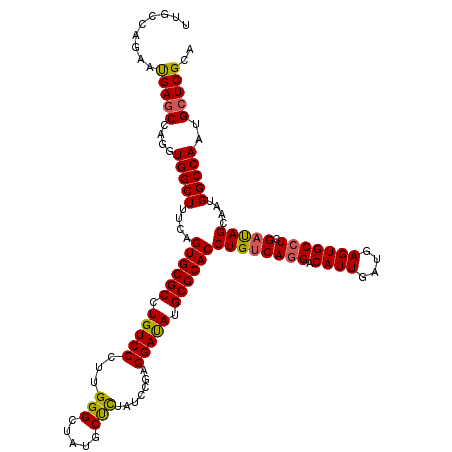
| Location | 7,540,453 – 7,540,573 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.33 |
| Mean single sequence MFE | -44.70 |
| Consensus MFE | -39.58 |
| Energy contribution | -39.22 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7540453 120 + 23771897 UUGCCAGAACGAGCCUGGUGGCUUUCAGUGCGCCUGUCCCUUGGGCUAUGCCCUAUCCGAGGAUAUGCGCACCUGUCAGGACAUUGAUGAGUGCCUCGAUAGCAAUGGCCAAUGCUCGCA .........(((((....(((((....((((((.((((((..((((...)))).....).))))).))))))((((((((.((((....))))))).)))))....)))))..))))).. ( -47.10) >DroSec_CAF1 9662 120 + 1 UUGUCAGAAUGAACCAGGUGGCUUUCAGUGCGCCUGUCCCUUGGGCCAUGCUCUAUCCGAGGAUAUGCGCACCUGUCAGGACAUUGACGAGUGCCUCGAUAGCAAUGGCCAAUGCUCGCA .(((.((.......((((((((.....)).)))))).......((((((...(((((.((((.....)((((.((((((....)))))).))))))))))))..))))))....)).))) ( -39.10) >DroSim_CAF1 9571 120 + 1 UUGUCAGAAUGAGCCAGGUGGCUUUCAGUGCGCCUGUCCCUUGGGCUAUGCUCUAUCCGAGGAUAUGCGCACCUGUCAGGACAUUGACGAGUGCCUCGAUAGCAAUGGCCAAUGCUCGCA ..........((((....(((((....((((((.(((((.(((((..........)))))))))).))))))((((((((.((((....))))))).)))))....)))))..))))... ( -44.10) >DroEre_CAF1 9779 120 + 1 UUGCCAGAAUGAGCCAGGUGGCUUCCUGUGCGCCUGUCCUUUGGGAUAUGCCUUAUCAGAGGACAUGCGCACCUGCCAGGACAUUGAUGAGUGCCUCGAUAGCAAUGGCCAGUGCUCGCA .(((.((..((.((((..((((.....((((((.((((((((((...........)))))))))).))))))..))))((.((((....))))))..........))))))...)).))) ( -45.90) >DroYak_CAF1 9667 120 + 1 UUGCCAGAAUGAGCCAGGUGGCUUCCAGUGCGCCUGUCCUUUGGGAUAUGCUUUAUCAGAGGAUAUGCGCACCUGUCAGGACAUUGAUGAGUGCAUUGACAGCAAUGGCCAAUGCUCGCA ..........((((....(((((....((((((.((((((((((...........)))))))))).))))))(((((((..((((....))))..)))))))....)))))..))))... ( -47.30) >consensus UUGCCAGAAUGAGCCAGGUGGCUUUCAGUGCGCCUGUCCCUUGGGCUAUGCUCUAUCCGAGGAUAUGCGCACCUGUCAGGACAUUGAUGAGUGCCUCGAUAGCAAUGGCCAAUGCUCGCA .........(((((....(((((....((((((.(((((...(((.....))).......))))).))))))((((((((.((((....))))))).)))))....)))))..))))).. (-39.58 = -39.22 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:43 2006