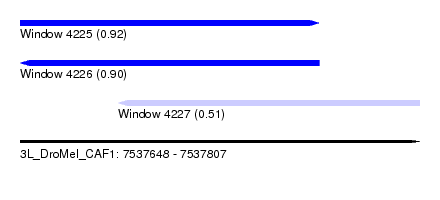
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,537,648 – 7,537,807 |
| Length | 159 |
| Max. P | 0.920062 |

| Location | 7,537,648 – 7,537,767 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.58 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
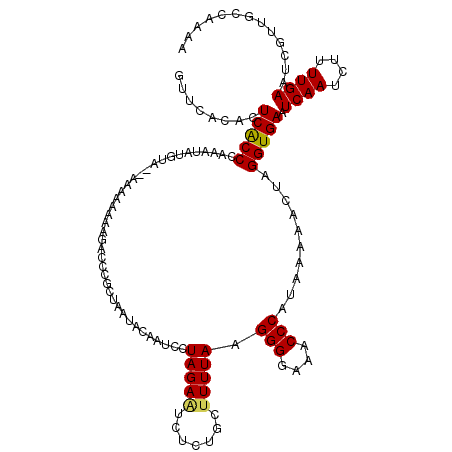
>3L_DroMel_CAF1 7537648 119 + 23771897 AAU-UAAAAAACCGUGAAGACUAUAUGGCAAAGAAAUAUCUUUUGGCAACGAUUCAAAAGAUUGAAUCACCUAGUUUUUAUGGGAUUCCCCUUAAAGGCAAAGAUUCUAGGAUAAUACAA ...-.......((((((((((((..((.((((((....)).)))).))..(((((((....)))))))...))))))))))))......(((..((........))..)))......... ( -26.50) >DroSec_CAF1 6830 120 + 1 AAUUUAAAAAACCGUGAAGACUAGAUGGCAAUAAUAUAACUUUUGGCAAAGAUUCAAAAGAUUGAUUCACCUAGUUUUUAUGGGUUUCCCCUUAAAAGCAGAGAUUCUAGAAUUGUAUUG (((((......(((((((((((((.((.(((...........))).))..((.((((....)))).))..)))))))))))))(((((..((....))..)))))....)))))...... ( -24.80) >DroSim_CAF1 6749 119 + 1 AAU-UAAAAAACCGUGAAGACUAGAUGGCAAUGAAAUAACUUUUGGCAACGAUUCAAAAGAUUGAUUCACCUAGUUUUUAUGGGUUUCCCCUUAAAAGCAGAGAUUCUAGGAUUGUAUUG ...-.......(((((((((((((.((.(((...........))).))..((.((((....)))).))..)))))))))))))(((((..((....))..)))))............... ( -24.60) >DroEre_CAF1 6929 100 + 1 AAU-U-------------------UUAGGAAUGAAAAAGCUUUUGGCAACGAUUCAAAAGAUUGAUUCACCUAGUUUUUAUGGGUUUCCCCGUAAAAGCUGAGACUCUAGGAUUGUAUUA (((-(-------------------((((((((.((....(((((((.......))))))).)).)))).(.(((((((((((((....))))))))))))).)...))))))))...... ( -27.50) >DroYak_CAF1 6889 100 + 1 AAU-U-------------------CUGGCCAUGAAAAAUCUUUUGGCAACGAUUCAAAAGAUUGAUUCACCUAGUUUUUAUGGGUUUCCCCUUAAAAGCUAAGAUUCUAGGAUUGUGUUA (((-(-------------------((((...((((.((((((((((.......))))))))))..))))(.(((((((((.(((....))).))))))))).)...))))))))...... ( -28.30) >consensus AAU_UAAAAAACCGUGAAGACUA_AUGGCAAUGAAAUAACUUUUGGCAACGAUUCAAAAGAUUGAUUCACCUAGUUUUUAUGGGUUUCCCCUUAAAAGCAGAGAUUCUAGGAUUGUAUUA ........................................((((((....((.((((....)))).)).....(((((((.(((....))).))))))).......))))))........ (-14.90 = -14.58 + -0.32)


| Location | 7,537,648 – 7,537,767 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -15.58 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.904196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7537648 119 - 23771897 UUGUAUUAUCCUAGAAUCUUUGCCUUUAAGGGGAAUCCCAUAAAAACUAGGUGAUUCAAUCUUUUGAAUCGUUGCCAAAAGAUAUUUCUUUGCCAUAUAGUCUUCACGGUUUUUUA-AUU ...........(((((.....(((((((.(((....))).)))).((((((((((((((....)))))))...))).(((((....)))))......))))......))).)))))-... ( -20.90) >DroSec_CAF1 6830 120 - 1 CAAUACAAUUCUAGAAUCUCUGCUUUUAAGGGGAAACCCAUAAAAACUAGGUGAAUCAAUCUUUUGAAUCUUUGCCAAAAGUUAUAUUAUUGCCAUCUAGUCUUCACGGUUUUUUAAAUU ...........(((((...(((.(((((.(((....))).)))))((((((((..((((....))))..((((....))))............)))))))).....)))..))))).... ( -23.70) >DroSim_CAF1 6749 119 - 1 CAAUACAAUCCUAGAAUCUCUGCUUUUAAGGGGAAACCCAUAAAAACUAGGUGAAUCAAUCUUUUGAAUCGUUGCCAAAAGUUAUUUCAUUGCCAUCUAGUCUUCACGGUUUUUUA-AUU ...........(((((...(((.(((((.(((....))).)))))((((((((...((((((((((.........)))))).......)))).)))))))).....)))..)))))-... ( -23.31) >DroEre_CAF1 6929 100 - 1 UAAUACAAUCCUAGAGUCUCAGCUUUUACGGGGAAACCCAUAAAAACUAGGUGAAUCAAUCUUUUGAAUCGUUGCCAAAAGCUUUUUCAUUCCUAA-------------------A-AUU .............(((....(((((((..(((....)))..........((..(.((((....))))....)..)))))))))..)))........-------------------.-... ( -22.20) >DroYak_CAF1 6889 100 - 1 UAACACAAUCCUAGAAUCUUAGCUUUUAAGGGGAAACCCAUAAAAACUAGGUGAAUCAAUCUUUUGAAUCGUUGCCAAAAGAUUUUUCAUGGCCAG-------------------A-AUU .........((......(((((.(((((.(((....))).))))).)))))((((..(((((((((.........))))))))).)))).))....-------------------.-... ( -24.30) >consensus UAAUACAAUCCUAGAAUCUCUGCUUUUAAGGGGAAACCCAUAAAAACUAGGUGAAUCAAUCUUUUGAAUCGUUGCCAAAAGAUAUUUCAUUGCCAU_UAGUCUUCACGGUUUUUUA_AUU .............(((.....((((((..(((....)))..........((..(.((((....))))....)..))))))))...)))................................ (-15.58 = -16.02 + 0.44)

| Location | 7,537,687 – 7,537,807 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.89 |
| Mean single sequence MFE | -19.30 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.30 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7537687 120 - 23771897 GUUCACAAUCACCCAAAUAUGUUUAAAAAAAAAAGCCCGAUUGUAUUAUCCUAGAAUCUUUGCCUUUAAGGGGAAUCCCAUAAAAACUAGGUGAUUCAAUCUUUUGAAUCGUUGCCAAAA ....((((((............................)))))).....(((((...(((.......)))(((...))).......))))).(((((((....))))))).......... ( -18.49) >DroSec_CAF1 6870 119 - 1 GUUCACACUCACCCAAAUAUGUU-AAAAAAAAAGACCCGCCAAUACAAUUCUAGAAUCUCUGCUUUUAAGGGGAAACCCAUAAAAACUAGGUGAAUCAAUCUUUUGAAUCUUUGCCAAAA .......................-......(((((..((((..........(((.....))).(((((.(((....))).)))))....))))..((((....)))).)))))....... ( -19.30) >DroSim_CAF1 6788 110 - 1 GUUCACCCUCACCCAA----------AAAAAAAGACCCGCCAAUACAAUCCUAGAAUCUCUGCUUUUAAGGGGAAACCCAUAAAAACUAGGUGAAUCAAUCUUUUGAAUCGUUGCCAAAA .((((((.........----------.........................(((.....))).(((((.(((....))).)))))....))))))((((....))))............. ( -18.50) >DroEre_CAF1 6949 118 - 1 GUUCACACUCACCCAAAAAUGUA--AAAAAAAAGAACCACUAAUACAAUCCUAGAGUCUCAGCUUUUACGGGGAAACCCAUAAAAACUAGGUGAAUCAAUCUUUUGAAUCGUUGCCAAAA ........(((((.......(((--(((....(((....(((.........)))..)))....))))))(((....)))..........))))).((((....))))............. ( -20.30) >DroYak_CAF1 6909 118 - 1 GUUCACUCUCGCCUUAAUAUGUA--CCAAAAAAUAAUGACUAACACAAUCCUAGAAUCUUAGCUUUUAAGGGGAAACCCAUAAAAACUAGGUGAAUCAAUCUUUUGAAUCGUUGCCAAAA .......................--...............((((.............(((((.(((((.(((....))).))))).))))).((.((((....)))).))))))...... ( -19.90) >consensus GUUCACACUCACCCAAAUAUGUA__AAAAAAAAGACCCGCUAAUACAAUCCUAGAAUCUCUGCUUUUAAGGGGAAACCCAUAAAAACUAGGUGAAUCAAUCUUUUGAAUCGUUGCCAAAA ........(((((......................................(((((.......))))).(((....)))..........))))).((((....))))............. (-14.38 = -14.30 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:39 2006