
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,536,351 – 7,536,603 |
| Length | 252 |
| Max. P | 0.998866 |

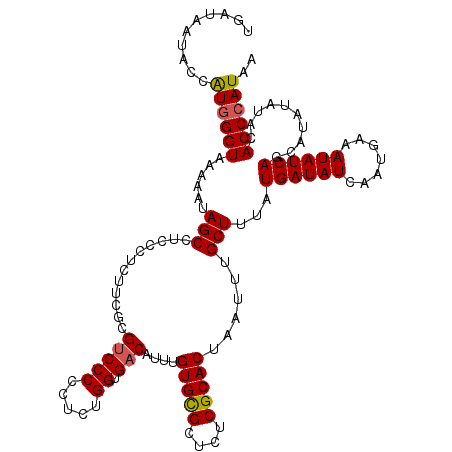
| Location | 7,536,351 – 7,536,471 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7536351 120 + 23771897 UGAUAAUACCAUGGGUAAAAAUAGCUUUCCUCUACGCGUCCCCCUCUGGUGACAUUUGUGCGCUCUCGCACUAAUUUGCUUUAUGAUAUCAAUGAAAUAUCAGCAUAUAUACACCCAUAA ..........((((((....(((((..........(((((((.....)).)))....(((((....)))))......))....((((((.......)))))))).)))....)))))).. ( -25.40) >DroSec_CAF1 5443 120 + 1 UGAUAAUACCAUGGGUAAAAAUAGCCUCCCCCUUCGCGUCCCCCUCUGGUGACAUUUGUGCGCUCUCGCACUAAUUUGCUUUAUGAUAUCUAUGAAAUAUCAGCAUAUAUACACCCAUAA ..........((((((....(((((..........(((((((.....)).)))....(((((....)))))......))....((((((.......)))))))).)))....)))))).. ( -25.00) >DroSim_CAF1 5385 120 + 1 UGAUAAUACCAUGGGUAAAAAUAGCCUCCCUCUUCGCGUCCCCCUCUGGUGACAUUUGUGCGCUCUCGCACUAAUUUGCUUUAUGAUAUCAAUGAAAUAUCAGCAUAUAUACACCCAUAA ..........((((((....(((((..........(((((((.....)).)))....(((((....)))))......))....((((((.......)))))))).)))....)))))).. ( -25.00) >DroEre_CAF1 5615 120 + 1 UGAUAAUACCGUGGGUAAAAAUAGCCUCCCUCUUUACGUCCCUCUCUGGUGACAUUUGUGUGCUCUCGCACUAAUUUGCUUUAUGAUAUCAAUGAAAUAUCAGCAUAUAUACACCCAUAA ..........((((((.....................(((((.....)).)))...(((((((....(((......)))....((((((.......)))))))))))))...)))))).. ( -26.60) >DroYak_CAF1 5577 120 + 1 UGAUAAUACCAUCGGUAAAAAUAGCCUCCCUCUUUACGCCCCCCUCUGGUGACAUUUGUGCGCUCUCGCACUAAUUUGCUUUAUGAUAUCAAUGAAAUAUCAGCAUAUAUACACCCAUAA (((((....(((.((((.....(((...........((((.......))))......(((((....)))))......)))......)))).)))...))))).................. ( -17.90) >consensus UGAUAAUACCAUGGGUAAAAAUAGCCUCCCUCUUCGCGUCCCCCUCUGGUGACAUUUGUGCGCUCUCGCACUAAUUUGCUUUAUGAUAUCAAUGAAAUAUCAGCAUAUAUACACCCAUAA ..........((((((......(((............(((((.....)).)))....(((((....)))))......)))...((((((.......))))))..........)))))).. (-21.52 = -21.60 + 0.08)

| Location | 7,536,351 – 7,536,471 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -32.04 |
| Energy contribution | -31.72 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7536351 120 - 23771897 UUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAAAGCAAAUUAGUGCGAGAGCGCACAAAUGUCACCAGAGGGGGACGCGUAGAGGAAAGCUAUUUUUACCCAUGGUAUUAUCA (((((((((...(((.(((((((((.....)))))).....((......((((....))))......(((.((....)).))))).........))))))...)))))))))........ ( -34.30) >DroSec_CAF1 5443 120 - 1 UUAUGGGUGUAUAUAUGCUGAUAUUUCAUAGAUAUCAUAAAGCAAAUUAGUGCGAGAGCGCACAAAUGUCACCAGAGGGGGACGCGAAGGGGGAGGCUAUUUUUACCCAUGGUAUUAUCA (((((((((...(((.(((((((((.....)))))).....((......((((....))))......(((.((....)).))))).........))))))...)))))))))........ ( -34.10) >DroSim_CAF1 5385 120 - 1 UUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAAAGCAAAUUAGUGCGAGAGCGCACAAAUGUCACCAGAGGGGGACGCGAAGAGGGAGGCUAUUUUUACCCAUGGUAUUAUCA (((((((((...(((.(((((((((.....)))))).....((......((((....))))......(((.((....)).))))).........))))))...)))))))))........ ( -34.30) >DroEre_CAF1 5615 120 - 1 UUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAAAGCAAAUUAGUGCGAGAGCACACAAAUGUCACCAGAGAGGGACGUAAAGAGGGAGGCUAUUUUUACCCACGGUAUUAUCA (..((((((...(((.(((((((((.....)))))).............((((....))))....(((((.((.....))))))).........))))))...))))))..)........ ( -30.90) >DroYak_CAF1 5577 120 - 1 UUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAAAGCAAAUUAGUGCGAGAGCGCACAAAUGUCACCAGAGGGGGGCGUAAAGAGGGAGGCUAUUUUUACCGAUGGUAUUAUCA ((((.((((...(((.(((((((((.....)))))).............((((....))))....(((((.((....)).))))).........))))))...)))).))))........ ( -27.80) >consensus UUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAAAGCAAAUUAGUGCGAGAGCGCACAAAUGUCACCAGAGGGGGACGCGAAGAGGGAGGCUAUUUUUACCCAUGGUAUUAUCA (((((((((...(((.(((((((((.....)))))).....((......((((....))))......(((.((.....))))))).........))))))...)))))))))........ (-32.04 = -31.72 + -0.32)


| Location | 7,536,391 – 7,536,489 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 92.52 |
| Mean single sequence MFE | -16.30 |
| Consensus MFE | -15.31 |
| Energy contribution | -15.12 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7536391 98 + 23771897 CCCCUCUGGUGACAUUUGUGCGCUCUCGCACUAAUUUGCUUUAUGAUAUCAAUGAAAUAUCAGCAUAUAUACACCCAUAAACUUAUAUAUAUAUAUAU .......((((......(((((....))))).....((((....(((((.......)))))))))......))))....................... ( -15.60) >DroSim_CAF1 5425 98 + 1 CCCCUCUGGUGACAUUUGUGCGCUCUCGCACUAAUUUGCUUUAUGAUAUCAAUGAAAUAUCAGCAUAUAUACACCCAUAAACGUAUAUAUAUACAUAU .......((((......(((((....))))).....((((....(((((.......)))))))))......)))).......((((....)))).... ( -17.10) >DroEre_CAF1 5655 98 + 1 CCUCUCUGGUGACAUUUGUGUGCUCUCGCACUAAUUUGCUUUAUGAUAUCAAUGAAAUAUCAGCAUAUAUACACCCAUAAACAUGCAGAUAUAUAUAU .......((((.....(((((((....(((......)))....((((((.......)))))))))))))..))))....................... ( -16.50) >DroYak_CAF1 5617 90 + 1 CCCCUCUGGUGACAUUUGUGCGCUCUCGCACUAAUUUGCUUUAUGAUAUCAAUGAAAUAUCAGCAUAUAUACACCCAUAAACG--------UAUAUAU ......((....))...(((((....))))).....((((....(((((.......)))))))))(((((((..........)--------)))))). ( -16.00) >consensus CCCCUCUGGUGACAUUUGUGCGCUCUCGCACUAAUUUGCUUUAUGAUAUCAAUGAAAUAUCAGCAUAUAUACACCCAUAAACGUAUAUAUAUAUAUAU .......((((......(((((....))))).....((((....(((((.......)))))))))......))))....................... (-15.31 = -15.12 + -0.19)

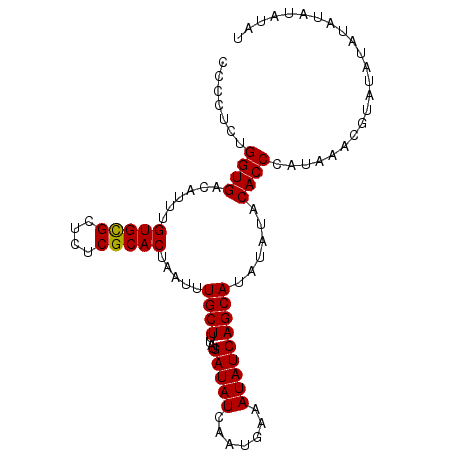
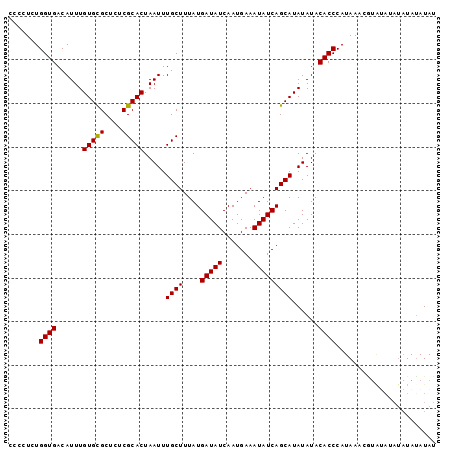
| Location | 7,536,391 – 7,536,489 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 92.52 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -21.89 |
| Energy contribution | -21.70 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7536391 98 - 23771897 AUAUAUAUAUAUAUAAGUUUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAAAGCAAAUUAGUGCGAGAGCGCACAAAUGUCACCAGAGGGG .(((((....)))))........((((.((((.((((((((((.....))))))....)))).....((((....))))....))))))))....... ( -21.90) >DroSim_CAF1 5425 98 - 1 AUAUGUAUAUAUAUACGUUUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAAAGCAAAUUAGUGCGAGAGCGCACAAAUGUCACCAGAGGGG ....((((((((((((.(....).))))))))))))(((((((.....)))))))............((((....))))..........((....)). ( -26.10) >DroEre_CAF1 5655 98 - 1 AUAUAUAUAUCUGCAUGUUUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAAAGCAAAUUAGUGCGAGAGCACACAAAUGUCACCAGAGAGG .(((((((((((..(....)..)))))))))))((((((((((.....))))))....)))).....((((....))))................... ( -22.90) >DroYak_CAF1 5617 90 - 1 AUAUAUA--------CGUUUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAAAGCAAAUUAGUGCGAGAGCGCACAAAUGUCACCAGAGGGG .......--------(.(((...((((.((((.((((((((((.....))))))....)))).....((((....))))....)))))))).))).). ( -22.30) >consensus AUAUAUAUAUAUAUACGUUUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAAAGCAAAUUAGUGCGAGAGCGCACAAAUGUCACCAGAGGGG .......................((((.((((.((((((((((.....))))))....)))).....((((....))))....))))))))....... (-21.89 = -21.70 + -0.19)

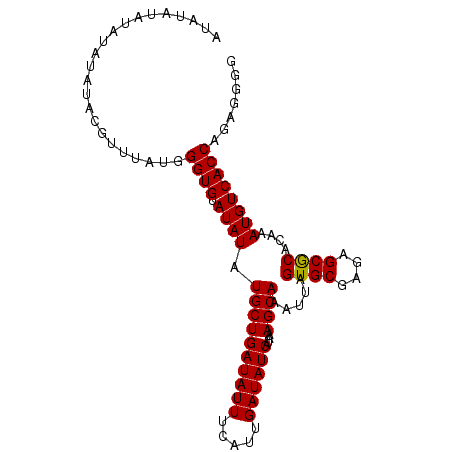
| Location | 7,536,431 – 7,536,523 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -19.75 |
| Consensus MFE | -13.01 |
| Energy contribution | -15.70 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7536431 92 - 23771897 AACGUGAAAACAGUUUUUCGAUGUCUCCAAAGAUAUAUAUAUAUAUAUAAGUUUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAA ...((....))........((((((.....((.(((((((((((.((((....)))).)))))))))))))((....))...)))))).... ( -17.70) >DroSim_CAF1 5465 92 - 1 AACGAGAAAACAGUUUUCCGAUGUCUCCAAAGAUAUAUGUAUAUAUAUACGUUUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAA ...(((((.....))))).((((((.....((((((..((((((((((((.(....).))))))))))))..))))))....)))))).... ( -23.40) >DroEre_CAF1 5695 91 - 1 AACGAGAA-ACAGUUUUUCGGUGUCUCCAAAGAUAUAUAUAUAUCUGCAUGUUUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAA ..((((((-.....))))))..........((.(((((((((((((..(....)..)))))))))))))))((((((.....)))))).... ( -19.40) >DroYak_CAF1 5657 84 - 1 AACGAGAAAACCAUUUUUCGAUGUCUCCAAAGAUAUAUAUA--------CGUUUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAA ...((((((....))))))((((((.......(((((((((--------(.(....).))))))))))..(((....)))..)))))).... ( -18.50) >consensus AACGAGAAAACAGUUUUUCGAUGUCUCCAAAGAUAUAUAUAUAUAUAUACGUUUAUGGGUGUAUAUAUGCUGAUAUUUCAUUGAUAUCAUAA ...((((((....))))))((((((.....((((((...(((((((((((.(....).)))))))))))...))))))....)))))).... (-13.01 = -15.70 + 2.69)

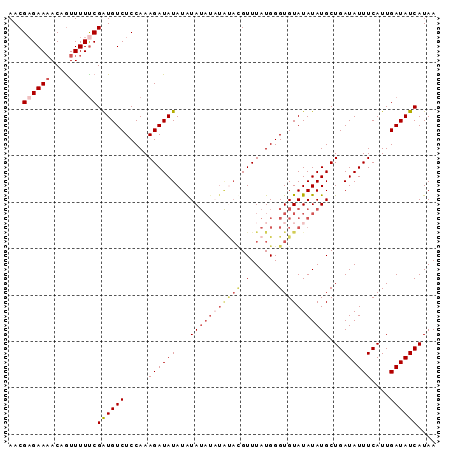
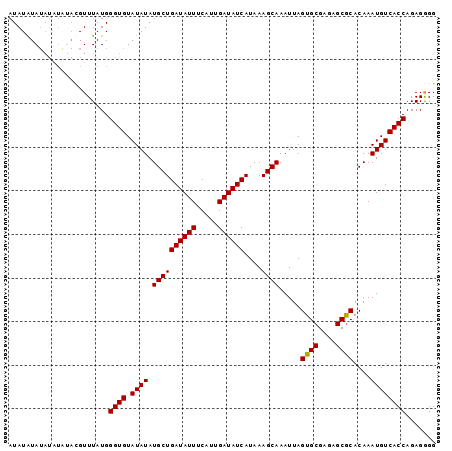
| Location | 7,536,471 – 7,536,563 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -20.02 |
| Consensus MFE | -19.15 |
| Energy contribution | -18.90 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7536471 92 + 23771897 ACUUAUAUAUAUAUAUAUAUCUUUGGAGACAUCGAAAAACUGUUUUCACGUUCCCUUCGAGAAAAGUCAGAAUUUCUUUGGCUCCUCAGUCU ...((((((....)))))).((..((((.((..((((..((((((((.((.......)).)))))..)))..))))..)).))))..))... ( -16.90) >DroSim_CAF1 5505 92 + 1 ACGUAUAUAUAUACAUAUAUCUUUGGAGACAUCGGAAAACUGUUUUCUCGUUCCCUUCGAGAAAAGUCAGAAUUUCUUUGGCUCCUCAGUCU ..((((....))))......((..((((.((..((((..(((((((((((.......))))))))..)))..))))..)).))))..))... ( -21.70) >DroEre_CAF1 5735 91 + 1 ACAUGCAGAUAUAUAUAUAUCUUUGGAGACACCGAAAAACUGU-UUCUCGUUCCCUUCGAGAAAAGUCAGAAUUUCUUUGGCUCCUCAGUCU ......((((((....))))))..((((.((..((((..((((-((((((.......)))))))...)))..))))..)).))))....... ( -21.00) >DroYak_CAF1 5697 84 + 1 ACG--------UAUAUAUAUCUUUGGAGACAUCGAAAAAUGGUUUUCUCGUUCCCUUCGAGAAAAGUCCGAAUUUCUUUGGCUCCUCAGUCU ...--------.........((..((((.((..((((..(((((((((((.......))))))))..)))..))))..)).))))..))... ( -20.50) >consensus ACGUAUAUAUAUAUAUAUAUCUUUGGAGACAUCGAAAAACUGUUUUCUCGUUCCCUUCGAGAAAAGUCAGAAUUUCUUUGGCUCCUCAGUCU ....................((..((((.((..((((..(((.(((((((.......)))))))...)))..))))..)).))))..))... (-19.15 = -18.90 + -0.25)


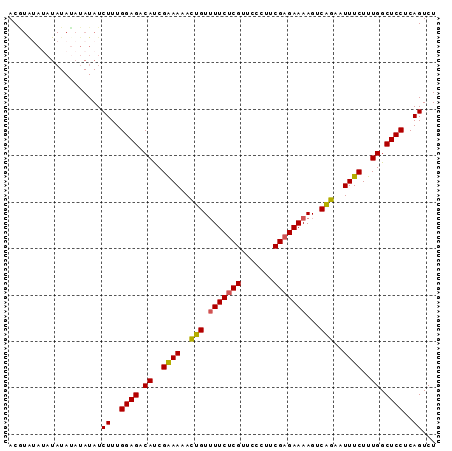
| Location | 7,536,471 – 7,536,563 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -19.70 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7536471 92 - 23771897 AGACUGAGGAGCCAAAGAAAUUCUGACUUUUCUCGAAGGGAACGUGAAAACAGUUUUUCGAUGUCUCCAAAGAUAUAUAUAUAUAUAUAAGU ...((..((((.((..((((..(((..(((((.((.......)).))))))))..))))..)).))))..))((((((....)))))).... ( -20.70) >DroSim_CAF1 5505 92 - 1 AGACUGAGGAGCCAAAGAAAUUCUGACUUUUCUCGAAGGGAACGAGAAAACAGUUUUCCGAUGUCUCCAAAGAUAUAUGUAUAUAUAUACGU ...((..((((.((..((((..(((..((((((((.......))))))))))).))))...)).))))..))....((((((....)))))) ( -21.50) >DroEre_CAF1 5735 91 - 1 AGACUGAGGAGCCAAAGAAAUUCUGACUUUUCUCGAAGGGAACGAGAA-ACAGUUUUUCGGUGUCUCCAAAGAUAUAUAUAUAUCUGCAUGU .......((((.((..((((..(((...(((((((.......))))))-))))..))))..)).))))..((((((....))))))...... ( -26.00) >DroYak_CAF1 5697 84 - 1 AGACUGAGGAGCCAAAGAAAUUCGGACUUUUCUCGAAGGGAACGAGAAAACCAUUUUUCGAUGUCUCCAAAGAUAUAUAUA--------CGU ...((..((((.((..((((...((..((((((((.......))))))))))...))))..)).))))..)).........--------... ( -20.60) >consensus AGACUGAGGAGCCAAAGAAAUUCUGACUUUUCUCGAAGGGAACGAGAAAACAGUUUUUCGAUGUCUCCAAAGAUAUAUAUAUAUAUAUACGU ...((..((((.((..((((..(((..((((((((.......)))))))))))..))))..)).))))..)).................... (-19.70 = -20.32 + 0.62)


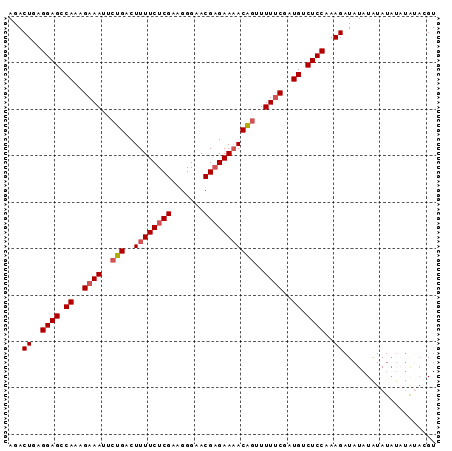
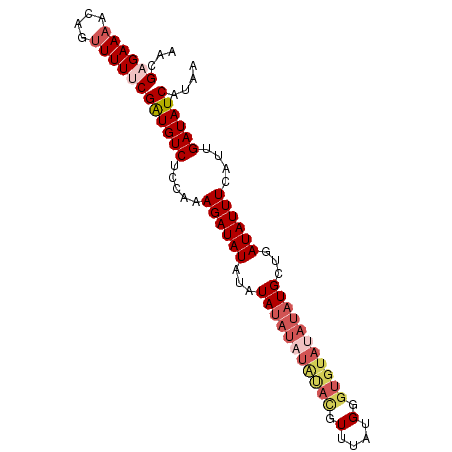
| Location | 7,536,489 – 7,536,603 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -29.92 |
| Energy contribution | -30.52 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7536489 114 + 23771897 ------AUCUUUGGAGACAUCGAAAAACUGUUUUCACGUUCCCUUCGAGAAAAGUCAGAAUUUCUUUGGCUCCUCAGUCUUGUCUGGGCUCUCGACGGAAUCGGUGCGUUUUCCAAAAUA ------...(((((((((((((((((....)))))..(((((..((((((..(((((((.....))))))).(((((......))))).)))))).))))).))))...))))))))... ( -34.30) >DroSec_CAF1 5603 120 + 1 AUAUAAAUCUUUGGAGACAUCGGAAAACUGUUUGCUCGUUUCCUUCGAGAAAAGUCCGAAUUUCUUUGGCUCAUCAGUCUUGUCUGGGCUCUCGACGGAAUCGGUGCGUUUUCCAAAAUA ...........((....))..(((((((.((..((.((.((((.((((((...(((((.((......((((....))))..)).))))))))))).)))).)))))))))))))...... ( -34.40) >DroSim_CAF1 5523 114 + 1 ------AUCUUUGGAGACAUCGGAAAACUGUUUUCUCGUUCCCUUCGAGAAAAGUCAGAAUUUCUUUGGCUCCUCAGUCUUGUCUGGGCUCUCGACGGAAUCGGUGCGUUUUCCAAAAUA ------.....((....))..(((((((.((...((.(((((..((((((..(((((((.....))))))).(((((......))))).)))))).))))).)).)))))))))...... ( -36.30) >DroEre_CAF1 5753 113 + 1 ------AUCUUUGGAGACACCGAAAAACUGU-UUCUCGUUCCCUUCGAGAAAAGUCAGAAUUUCUUUGGCUCCUCAGUCUUUCCUGGCCUCUCGACGGAACCGGUGCGUUUUCCAAAAUA ------...((((((((((((((((.....)-)))..(((((..((((((..(((((((.....))))))).....(((......))).)))))).))))).))))...))))))))... ( -34.50) >DroYak_CAF1 5707 114 + 1 ------AUCUUUGGAGACAUCGAAAAAUGGUUUUCUCGUUCCCUUCGAGAAAAGUCCGAAUUUCUUUGGCUCCUCAGUCUUGCCUGGGCUCUCGACGGAAUCGGUGCGUUUUCCAAAAUA ------...((((((((((((((..(((((.....))))).((.((((((..((.(((((....))))))).(((((......))))).)))))).))..))))))...))))))))... ( -32.20) >consensus ______AUCUUUGGAGACAUCGAAAAACUGUUUUCUCGUUCCCUUCGAGAAAAGUCAGAAUUUCUUUGGCUCCUCAGUCUUGUCUGGGCUCUCGACGGAAUCGGUGCGUUUUCCAAAAUA .........(((((((((((((((((....)))))..(((((..((((((..(((((((.....))))))).(((((......))))).)))))).))))).))))...))))))))... (-29.92 = -30.52 + 0.60)


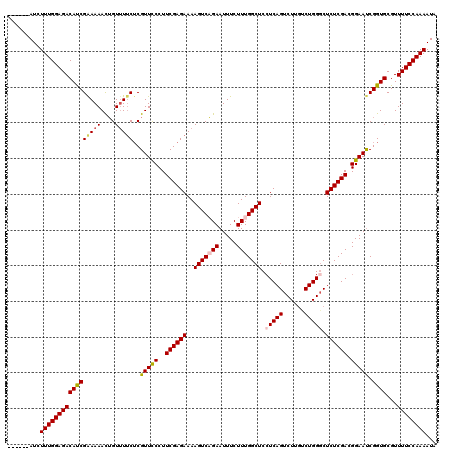
| Location | 7,536,489 – 7,536,603 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -23.92 |
| Energy contribution | -23.56 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7536489 114 - 23771897 UAUUUUGGAAAACGCACCGAUUCCGUCGAGAGCCCAGACAAGACUGAGGAGCCAAAGAAAUUCUGACUUUUCUCGAAGGGAACGUGAAAACAGUUUUUCGAUGUCUCCAAAGAU------ ..(((((((....(((.(((((((.((((((((((((......))).))((.((((....)).)).)).))))))).))))..((....))......))).))).)))))))..------ ( -28.40) >DroSec_CAF1 5603 120 - 1 UAUUUUGGAAAACGCACCGAUUCCGUCGAGAGCCCAGACAAGACUGAUGAGCCAAAGAAAUUCGGACUUUUCUCGAAGGAAACGAGCAAACAGUUUUCCGAUGUCUCCAAAGAUUUAUAU ....(((((((((((..((.((((.(((((((..(((......)))..(((((.((....)).)).)))))))))).)))).)).)).....))))))))).((((....))))...... ( -30.50) >DroSim_CAF1 5523 114 - 1 UAUUUUGGAAAACGCACCGAUUCCGUCGAGAGCCCAGACAAGACUGAGGAGCCAAAGAAAUUCUGACUUUUCUCGAAGGGAACGAGAAAACAGUUUUCCGAUGUCUCCAAAGAU------ ....(((((((((...(((.((((.((((((((((((......))).))((.((((....)).)).)).))))))).)))).)).)......))))))))).((((....))))------ ( -28.00) >DroEre_CAF1 5753 113 - 1 UAUUUUGGAAAACGCACCGGUUCCGUCGAGAGGCCAGGAAAGACUGAGGAGCCAAAGAAAUUCUGACUUUUCUCGAAGGGAACGAGAA-ACAGUUUUUCGGUGUCUCCAAAGAU------ ..(((((((....((((((((((((((....)))(((......))).))))))...((((..(((...(((((((.......))))))-))))..))))))))).)))))))..------ ( -44.20) >DroYak_CAF1 5707 114 - 1 UAUUUUGGAAAACGCACCGAUUCCGUCGAGAGCCCAGGCAAGACUGAGGAGCCAAAGAAAUUCGGACUUUUCUCGAAGGGAACGAGAAAACCAUUUUUCGAUGUCUCCAAAGAU------ ..(((((((...........((((.((((((((((((......))).))((((.((....)).)).)).))))))).)))).(((((((....))))))).....)))))))..------ ( -32.50) >consensus UAUUUUGGAAAACGCACCGAUUCCGUCGAGAGCCCAGACAAGACUGAGGAGCCAAAGAAAUUCUGACUUUUCUCGAAGGGAACGAGAAAACAGUUUUUCGAUGUCUCCAAAGAU______ ....(((((((((...(((.((((.(((((((..(((......)))..((((..((....))..).)))))))))).)))).)).)......))))))))).((((....))))...... (-23.92 = -23.56 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:36 2006