
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,533,678 – 7,533,776 |
| Length | 98 |
| Max. P | 0.890171 |

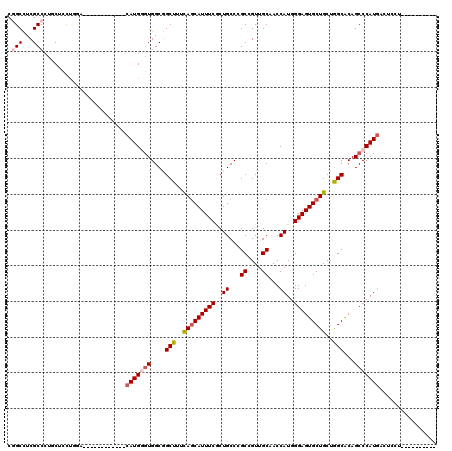
| Location | 7,533,678 – 7,533,776 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.55 |
| Mean single sequence MFE | -41.85 |
| Consensus MFE | -25.29 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7533678 98 + 23771897 CGGCCUCGCCCUUCUGUUUGA------------CAUGGCUGGCGGCUUUCAGCAUUUCGCUGCCCGCCGUUGCAACCAUGGGAGUGCUGCUGGCACAGCCCAUGACUUCU---------- .(((...))).......(((.------------.(((((.((((((............)))))).)))))..))).((((((.((((.....))))..))))))......---------- ( -36.10) >DroPse_CAF1 4266 108 + 1 CGGCCUCGCCCUUCUCCCGGA------------CAUGGGUGGCGGCUUUCAGCAUUUCGCUGCCGGCCGUUGCAACCAUGGGAGUGCUGCUGGCACAGCCCAUGACUCCUGUGGGGCUGC (((((((((((.......)).------------...(((..((((((..((((.....))))..))))))..)...((((((.((((.....))))..))))))...)).))))))))). ( -51.80) >DroGri_CAF1 4342 108 + 1 CGGCUUCGCCUUGCUUCUUGACAUCGCUUGCCACAUGAGUGGCGGCUUUUAGCAUUUCACUGCCUGCAGUUGCCACCAUUGGAGUGCUGCUAGCACAGCCCAUUGCUA------------ .((((..((..(((.....).))..)).((((((....))))))(((..((((((((((.((...((....))...)).))))))))))..)))..))))........------------ ( -34.60) >DroWil_CAF1 11754 110 + 1 CCGCCUCGCCUUGCUGCUUAACAUCACCAGCCACAUGGGUGGCAGCUUUCAACAUUUCACUGCCCGCAGUUGCCACCAUGGGAGUGCUGCUGGCGCAGCCCAUGACUUUU---------- ............((((((((.((.(((...((.(((((.((((((((.....((......)).....))))))))))))))).))).)).))).)))))...........---------- ( -33.90) >DroMoj_CAF1 4787 116 + 1 CCGCUUCGCCUUGCUUUUGAACAUCGCCUGCCACAUGUGUGGCAGCUUUUAGCAUUUCGCUGCCAGCCGUUGCAACCAUCGGAGUGCUGCUGGCGCAGCCCAUGGCUGCUCUC----UCU .......(((..((...........((.((((((....))))))))....(((((((((.((...((....))...)).))))))))))).)))((((((...))))))....----... ( -42.90) >DroPer_CAF1 4315 108 + 1 CGGCCUCGCCCUUCUCCCGGA------------CAUGGGUGGCGGCUUUCAGCAUUUCGCUGCCGGCCGUUGCAACCAUGGGAGUGCUGCUGGCACAGCCCAUGACUCCUGUGGGGCUGC (((((((((((.......)).------------...(((..((((((..((((.....))))..))))))..)...((((((.((((.....))))..))))))...)).))))))))). ( -51.80) >consensus CGGCCUCGCCCUGCUCCUGGA____________CAUGGGUGGCGGCUUUCAGCAUUUCGCUGCCCGCCGUUGCAACCAUGGGAGUGCUGCUGGCACAGCCCAUGACUCCU__________ .(((...))).......................(((((((....(((..(((((((((..((...((....))...))..)))))))))..)))...)))))))................ (-25.29 = -26.10 + 0.81)


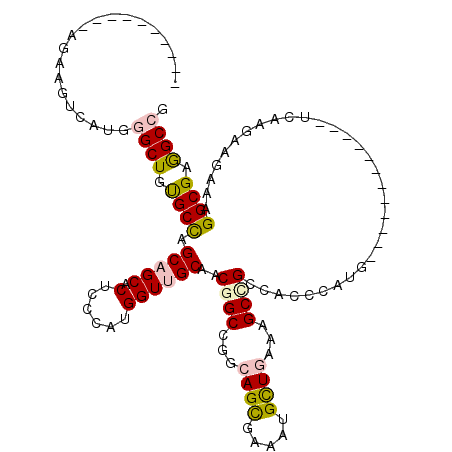
| Location | 7,533,678 – 7,533,776 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.55 |
| Mean single sequence MFE | -42.55 |
| Consensus MFE | -25.38 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7533678 98 - 23771897 ----------AGAAGUCAUGGGCUGUGCCAGCAGCACUCCCAUGGUUGCAACGGCGGGCAGCGAAAUGCUGAAAGCCGCCAGCCAUG------------UCAAACAGAAGGGCGAGGCCG ----------.(.(..((((((..((((.....)))).))))))..).)...(((...((((.....))))...)))(((.(((...------------((.....))..)))..))).. ( -39.40) >DroPse_CAF1 4266 108 - 1 GCAGCCCCACAGGAGUCAUGGGCUGUGCCAGCAGCACUCCCAUGGUUGCAACGGCCGGCAGCGAAAUGCUGAAAGCCGCCACCCAUG------------UCCGGGAGAAGGGCGAGGCCG (((((((....))...((((((..((((.....)))).)))))))))))..(((((..((((.....)))).....((((.(((...------------...))).....)))).))))) ( -46.40) >DroGri_CAF1 4342 108 - 1 ------------UAGCAAUGGGCUGUGCUAGCAGCACUCCAAUGGUGGCAACUGCAGGCAGUGAAAUGCUAAAAGCCGCCACUCAUGUGGCAAGCGAUGUCAAGAAGCAAGGCGAAGCCG ------------........((((....(((((.((((((..((((....))))..)).))))...)))))...((((((((....)))))..((...........))..)))..)))). ( -37.00) >DroWil_CAF1 11754 110 - 1 ----------AAAAGUCAUGGGCUGCGCCAGCAGCACUCCCAUGGUGGCAACUGCGGGCAGUGAAAUGUUGAAAGCUGCCACCCAUGUGGCUGGUGAUGUUAAGCAGCAAGGCGAGGCGG ----------....(((....(((((...((((.((((((((((((((((.((...((((......))))...)).))))).))))).))..)))).))))..)))))..)))....... ( -40.50) >DroMoj_CAF1 4787 116 - 1 AGA----GAGAGCAGCCAUGGGCUGCGCCAGCAGCACUCCGAUGGUUGCAACGGCUGGCAGCGAAAUGCUAAAAGCUGCCACACAUGUGGCAGGCGAUGUUCAAAAGCAAGGCGAAGCGG ...----....(((((((((((((((....)))))...)).))))))))..(.(((((((......)))).....(((((((....)))))))((..(((......)))..))..))).) ( -45.60) >DroPer_CAF1 4315 108 - 1 GCAGCCCCACAGGAGUCAUGGGCUGUGCCAGCAGCACUCCCAUGGUUGCAACGGCCGGCAGCGAAAUGCUGAAAGCCGCCACCCAUG------------UCCGGGAGAAGGGCGAGGCCG (((((((....))...((((((..((((.....)))).)))))))))))..(((((..((((.....)))).....((((.(((...------------...))).....)))).))))) ( -46.40) >consensus __________AGAAGUCAUGGGCUGUGCCAGCAGCACUCCCAUGGUUGCAACGGCCGGCAGCGAAAUGCUGAAAGCCGCCACCCAUG____________UCAAGAAGAAAGGCGAGGCCG ....................((((.((((.(((((.(......))))))..((((...((((.....))))...))))................................)))).)))). (-25.38 = -25.68 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:25 2006