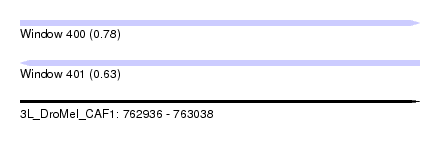
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 762,936 – 763,038 |
| Length | 102 |
| Max. P | 0.778480 |

| Location | 762,936 – 763,038 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -18.49 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
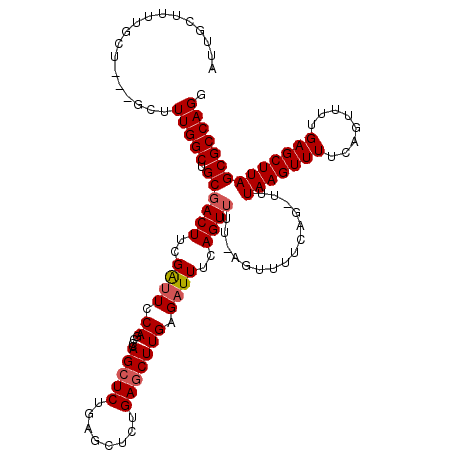
>3L_DroMel_CAF1 762936 102 + 23771897 AUUGCUUUUGCU---GCUUUGGCUGCAACUUCGAUUCCAGCCCAGCUCUGAGCUCUGAGCUUGAGAUUUCAGU--------------UUUAAGUUUUCAGUUUUGAGCUUAGCGCCAGG ....((..((((---(....(((((.((......)).))))).(((((.(((((..(((((((((((....))--------------)))))))))..))))).))))))))))..)). ( -32.20) >DroSim_CAF1 120097 116 + 1 AUUGCUUUUGCU---GCUUUGGCUGCGACUUCGAUUCCAGCCCAGCUCUGAGCUCUGAGCUUGAGAUUUCAGUUUUAAGUUUUCAGUUUUAAGUUUUCAGUUUUGAGCUUAGCGCCAGG .........(((---(....(((((.((.......))))))))))).(((.((.(((((((..(((((.....((((((........)))))).....)))))..))))))).))))). ( -36.20) >DroEre_CAF1 124023 112 + 1 UUUGCUGUGACUAUGGCUUUGGCUGCGACUUUGGUUGCAACCUAGGUCU-------GAGCUUGAGUUUUCAGUUUUCAGUUUUCAGCUUUAAGUUUUCAGUUUUGAGCUUAGCGCCAGG ...((((.((((..(((((.((((..(((((.(((....))).))))).-------.)))).)))))...))))..)))).....((.((((((((........)))))))).)).... ( -31.60) >consensus AUUGCUUUUGCU___GCUUUGGCUGCGACUUCGAUUCCAGCCCAGCUCUGAGCUCUGAGCUUGAGAUUUCAGUUUU_AGUUUUCAG_UUUAAGUUUUCAGUUUUGAGCUUAGCGCCAGG ..................(((((.((((((..((((.((....(((((........))))))).))))..))))...............(((((((........)))))))))))))). (-18.49 = -19.27 + 0.78)

| Location | 762,936 – 763,038 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -15.87 |
| Energy contribution | -15.77 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
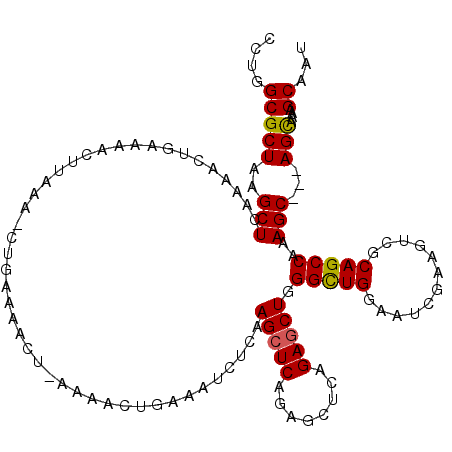
>3L_DroMel_CAF1 762936 102 - 23771897 CCUGGCGCUAAGCUCAAAACUGAAAACUUAAA--------------ACUGAAAUCUCAAGCUCAGAGCUCAGAGCUGGGCUGGAAUCGAAGUUGCAGCCAAAGC---AGCAAAAGCAAU .(((((.((.((((((....))).........--------------..(((....))).))).)).)).))).(((.(((((.(((....))).)))))..)))---.((....))... ( -25.80) >DroSim_CAF1 120097 116 - 1 CCUGGCGCUAAGCUCAAAACUGAAAACUUAAAACUGAAAACUUAAAACUGAAAUCUCAAGCUCAGAGCUCAGAGCUGGGCUGGAAUCGAAGUCGCAGCCAAAGC---AGCAAAAGCAAU .(((((.((.((((((....))).........................(((....))).))).)).)).))).(((.(((((((.......)).)))))..)))---.((....))... ( -24.20) >DroEre_CAF1 124023 112 - 1 CCUGGCGCUAAGCUCAAAACUGAAAACUUAAAGCUGAAAACUGAAAACUGAAAACUCAAGCUC-------AGACCUAGGUUGCAACCAAAGUCGCAGCCAAAGCCAUAGUCACAGCAAA ..((((.....)).))................((((...((((....((((..........))-------)).....((((((.((....)).)))))).......))))..))))... ( -22.30) >consensus CCUGGCGCUAAGCUCAAAACUGAAAACUUAAA_CUGAAAACU_AAAACUGAAAUCUCAAGCUCAGAGCUCAGAGCUGGGCUGGAAUCGAAGUCGCAGCCAAAGC___AGCAAAAGCAAU ....(((((..(((............................................(((((........))))).(((((............)))))..)))...)))....))... (-15.87 = -15.77 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:53 2006