| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,451,732 – 7,451,823 |
| Length | 91 |
| Max. P | 0.958821 |

| Location | 7,451,732 – 7,451,823 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.94 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -13.90 |
| Energy contribution | -14.48 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
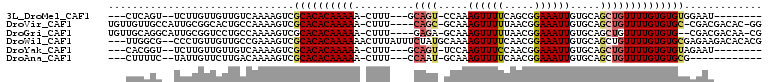
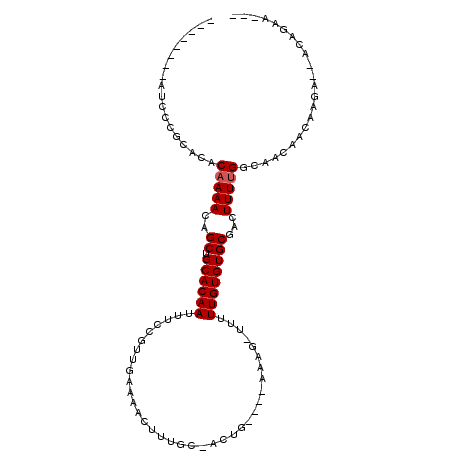
>3L_DroMel_CAF1 7451732 91 + 23771897 ---CUCAGU--UCUUGUUGUUGUCAAAAGUCGCACACAAAAA-CUUU---GCAGU-CCAAAGUUUUCAGCGGAAAUUGUGCAGCUGUUUUGUGUGUGGAAU-------- ---......--..................(((((((((((((-((((---(....-.))))))...((((............))))))))))))))))...-------- ( -22.20) >DroVir_CAF1 120902 101 + 1 UGUUGUUGCCAUUGCGGCACUGCCAAAAGUCGCACACAAAAA-CUUU----CAGC-GCAAAGUUUUUAACGGAAAUUGUGCAGCUGUUUUGUGUGC-CGACGACAC-GG ((((((((.......(((...))).......((((((((((.-....----((((-(((.((((((.....)))))).))).))))))))))))))-)))))))).-.. ( -34.50) >DroGri_CAF1 95871 100 + 1 UGUUGCAGGCAUUGCGGUCCUGCCAAAAGUCGCACACAAAAA-CUUU----GAGA-GCAAAGUUUUUAACGGAAAUUGUGCAGCUGUUUUGUGUG--CGACGACAA-CG ((((((((((......).))))).....((((((((((((((-((((----(...-.))))))))))(((((...........)))))..)))))--)))))))).-.. ( -36.20) >DroWil_CAF1 131261 104 + 1 ---UUGGCG--CCCUGUUGUUGCCGAAAGUCGCACACAAAAAACUUUAUUUCUAUGCAAAAGUUUUCAACGGAAAUUGUGCAGCUGUUUUGUGUGCGAGAAGACACACG ---((((((--(......).))))))...((((((((((((.............((((..((((((.....)))))).))))....))))))))))))........... ( -27.63) >DroYak_CAF1 99812 91 + 1 ---CACGGU--UCUUGUUGUUGUCAAAAGUCGCACACAAAAA-CUUU---GCAGU-UCCAAGUUUCCAACGGAAAUUGUGCAGCUGUUUUGUGUGUAGAAU-------- ---....((--(((..(((....))).....(((((((((..-....---(((((-(.(((((((((...))))))).)).))))))))))))))))))))-------- ( -23.10) >DroAna_CAF1 103720 87 + 1 ---CUUUUC--UAUUGUUCUUGACAAAAGUCGCACACAAAAA-CUUU---CCAAU-GCAAAGUUUUCAACGGAAAUUGUGCAGCUGUUUUGUGUGCG------------ ---......--..........(((....)))((((((((((.-....---....(-(((.((((((.....)))))).))))....)))))))))).------------ ( -23.62) >consensus ___CUCAGC__UCUUGUUGUUGCCAAAAGUCGCACACAAAAA_CUUU____CAGU_GCAAAGUUUUCAACGGAAAUUGUGCAGCUGUUUUGUGUGCGCGAC________ ...............................((((((((((..........((((.....((((((.....)))))).....))))))))))))))............. (-13.90 = -14.48 + 0.58)
| Location | 7,451,732 – 7,451,823 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 71.94 |
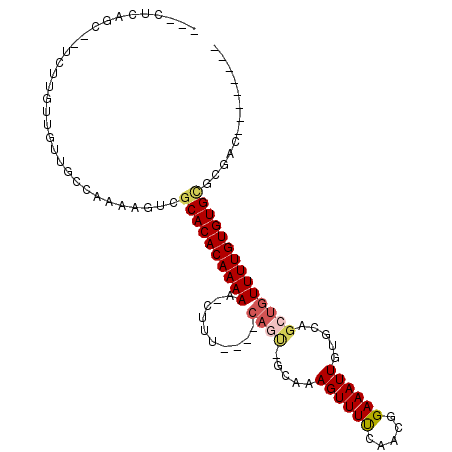
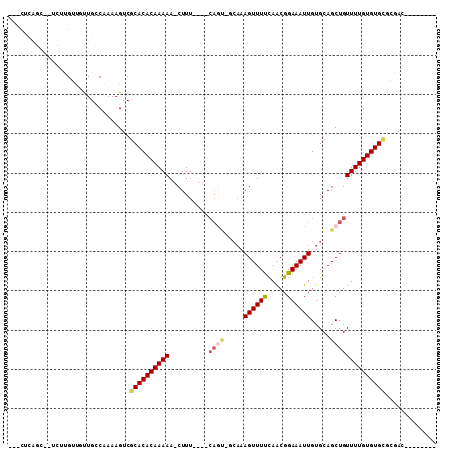
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -7.87 |
| Energy contribution | -8.04 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7451732 91 - 23771897 --------AUUCCACACACAAAACAGCUGCACAAUUUCCGCUGAAAACUUUGG-ACUGC---AAAG-UUUUUGUGUGCGACUUUUGACAACAACAAGA--ACUGAG--- --------....(.((((((...((((............))))(((((((((.-....)---))))-)))))))))).)..(((((.......)))))--......--- ( -19.50) >DroVir_CAF1 120902 101 - 1 CC-GUGUCGUCG-GCACACAAAACAGCUGCACAAUUUCCGUUAAAAACUUUGC-GCUG----AAAG-UUUUUGUGUGCGACUUUUGGCAGUGCCGCAAUGGCAACAACA ..-.((((((((-((((.((((..((.((((((........((((((((((..-....----))))-)))))))))))).))))))...))))))..))))))...... ( -30.50) >DroGri_CAF1 95871 100 - 1 CG-UUGUCGUCG--CACACAAAACAGCUGCACAAUUUCCGUUAAAAACUUUGC-UCUC----AAAG-UUUUUGUGUGCGACUUUUGGCAGGACCGCAAUGCCUGCAACA .(-((((.((((--(((((.....(((............)))((((((((((.-...)----))))-))))))))))))))....((((.........)))).))))). ( -32.60) >DroWil_CAF1 131261 104 - 1 CGUGUGUCUUCUCGCACACAAAACAGCUGCACAAUUUCCGUUGAAAACUUUUGCAUAGAAAUAAAGUUUUUUGUGUGCGACUUUCGGCAACAACAGGG--CGCCAA--- .(.(((((((.((((((((((((.(((((((.((((((....))))..)).))).((....)).)))))))))))))))).....(....)...))))--))))..--- ( -30.00) >DroYak_CAF1 99812 91 - 1 --------AUUCUACACACAAAACAGCUGCACAAUUUCCGUUGGAAACUUGGA-ACUGC---AAAG-UUUUUGUGUGCGACUUUUGACAACAACAAGA--ACCGUG--- --------......(((((((((....((((....(((((..(....).))))-).)))---)...-.)))))))))((..(((((.......)))))--..))..--- ( -20.90) >DroAna_CAF1 103720 87 - 1 ------------CGCACACAAAACAGCUGCACAAUUUCCGUUGAAAACUUUGC-AUUGG---AAAG-UUUUUGUGUGCGACUUUUGUCAAGAACAAUA--GAAAAG--- ------------((((((((...((((............))))((((((((.(-...).---))))-)))))))))))).((.((((.....)))).)--).....--- ( -21.10) >consensus ________AUCCCGCACACAAAACAGCUGCACAAUUUCCGUUGAAAACUUUGC_ACUG____AAAG_UUUUUGUGUGCGACUUUUGGCAACAACAAGA__ACAGAA___ ..................(((((..((.((((((....................................))))))))...)))))....................... ( -7.87 = -8.04 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:05 2006