
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,437,152 – 7,437,268 |
| Length | 116 |
| Max. P | 0.746210 |

| Location | 7,437,152 – 7,437,248 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.96 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -15.68 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
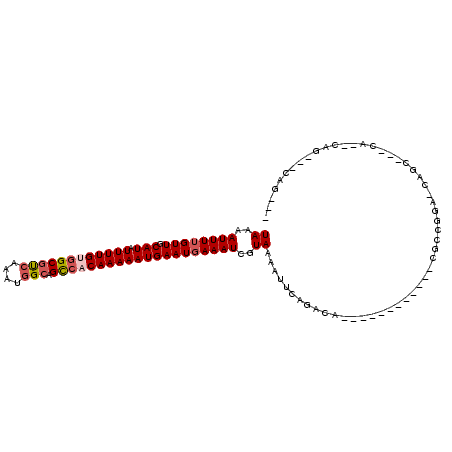
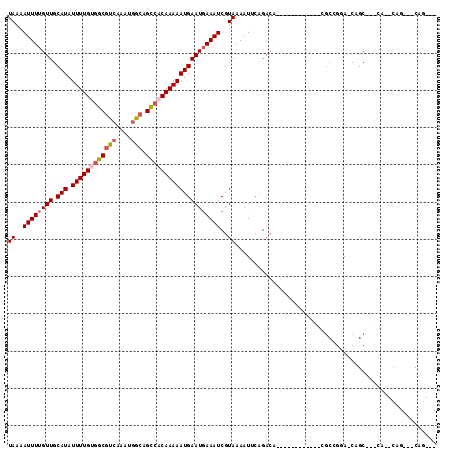
>3L_DroMel_CAF1 7437152 96 - 23771897 UAAAAUUUAGUUGCAUAUUUUGCGGCGCCAAAUGGCAGCCACAAAAAUGAAUGAAAUCGUAAAAUUCAGACC-----------CUGCUGGA-CCGCAUCCA--CGCC--CAG--- ((..((((.(((.(((.(((((.((((((....))).))).))))))))))).))))..))...........-----------..((((((-.....))))--.)).--...--- ( -21.10) >DroVir_CAF1 101822 91 - 1 UAAAAUUUUGUUGCAUAUUUUGUGGCGUCAAAUGGCAGCCACAAAAAUGAAUGAAAUCGUAAAAUCCAGACA------------CGCCGGA-CAGC---CA--CAG---CAG--- ((..((((..((.(((.((((((((((((....))).))))))))))))))..))))..))...(((.(...------------..).)))-..((---..--..)---)..--- ( -22.30) >DroGri_CAF1 81150 91 - 1 UAAAAUUUUGUUGCAUAUUUUGUGGCGUAAAAUAGCAGCCACAAAAAUGAAUGAAAUCGUAAAAUUCAGACA------------CGCCGGA-CAGC---CA--CAG---CAG--- ((..((((..((.(((.(((((((((((......)).))))))))))))))..))))..))...........------------.((.(..-....---..--).)---)..--- ( -19.50) >DroWil_CAF1 111470 110 - 1 UAAAAUUUUGUUGCAUAUUUUGUGGCAUCAAAUGGCAGCCACAAAAAUGAAUGAAAUCGUAAAAUUCGGACAAGCAAGCGAAGCCGCUGCAGCAGCCGCAA--CGCCAGCAG--- ((..((((..((.(((.(((((((((...........))))))))))))))..))))..))............((..(((..((.((((...)))).))..--)))..))..--- ( -30.70) >DroMoj_CAF1 94938 96 - 1 UAAAAUUUUGUUGCAUAUUUUGCGGCGUCGAAUGGCAGCCACAAAAAUGAAUGAAAUCGUAAAAUGCAGACA------------CGCCGGA-CAGC---CAGCCAG---CCGGCC ........((((((((.(((((.((((((....))).))).)))))((((......))))...))))).)))------------.(((((.-..(.---...)...---))))). ( -26.80) >DroAna_CAF1 88554 90 - 1 UAAAAUUUUGUUGCAUAUUUUGUGGCGUCGAAUGGCAGUAACAAAAAUGAAUGAAAUCGUAAAAUUUGGACC-----------GCACUGGA-CCGCAUC----------CAG--- ((..((((..((.(((.((((((.(((((....))).)).)))))))))))..))))..)).....((((..-----------((......-..)).))----------)).--- ( -16.70) >consensus UAAAAUUUUGUUGCAUAUUUUGUGGCGUCAAAUGGCAGCCACAAAAAUGAAUGAAAUCGUAAAAUUCAGACA____________CGCCGGA_CAGC___CA__CAG___CAG___ ((..((((((((.(((.((((((((((((....))).))))))))))))))))))))..))...................................................... (-15.68 = -16.40 + 0.72)

| Location | 7,437,176 – 7,437,268 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -20.98 |
| Consensus MFE | -15.19 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
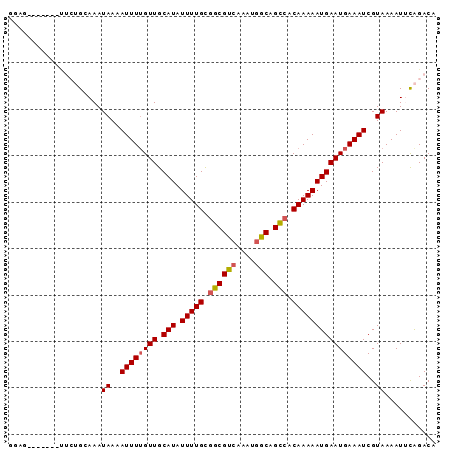
>3L_DroMel_CAF1 7437176 92 - 23771897 UGAGCUCUUAUUUCUGCAAAUAAAAUUUAGUUGCAUAUUUUGCGGCGCCAAAUGGCAGCCACAAAAAUGAAUGAAAUCGUAAAAUUCAGACC .((..((....((((((((.((.....)).)))))..(((((.((((((....))).))).)))))..))).))..)).............. ( -19.40) >DroVir_CAF1 101841 85 - 1 GGAG-------CUCUGGAAAUAAAAUUUUGUUGCAUAUUUUGUGGCGUCAAAUGGCAGCCACAAAAAUGAAUGAAAUCGUAAAAUCCAGACA ...(-------.((((((..((..((((..((.(((.((((((((((((....))).))))))))))))))..))))..))...))))))). ( -28.90) >DroGri_CAF1 81169 85 - 1 GGAG-------UUCUGGAAAUAAAAUUUUGUUGCAUAUUUUGUGGCGUAAAAUAGCAGCCACAAAAAUGAAUGAAAUCGUAAAAUUCAGACA ...(-------(.(((((..((..((((..((.(((.(((((((((((......)).))))))))))))))..))))..))...))))))). ( -23.40) >DroYak_CAF1 85046 92 - 1 CGUGCUUUUAUUUCGGCAAAUAAAAUUUUGUUGCAUAUUUUGCGGCGCCAAAUGGCAGCCACAAAAAUGAAUGAAAUCGUAAAAUUCAGACC ..((((........))))..((..((((..((.(((.(((((.((((((....))).))).))))))))))..))))..))........... ( -22.00) >DroMoj_CAF1 94962 83 - 1 GAAG-------C--UGGAAAUAAAAUUUUGUUGCAUAUUUUGCGGCGUCGAAUGGCAGCCACAAAAAUGAAUGAAAUCGUAAAAUGCAGACA ...(-------(--((.(..((..((((..((.(((.(((((.((((((....))).))).))))))))))..))))..))...).))).). ( -18.40) >DroAna_CAF1 88572 80 - 1 CGG------------GCAAAUAAAAUUUUGUUGCAUAUUUUGUGGCGUCGAAUGGCAGUAACAAAAAUGAAUGAAAUCGUAAAAUUUGGACC .((------------.((((((..((((..((.(((.((((((.(((((....))).)).)))))))))))..))))..))...))))..)) ( -13.80) >consensus GGAG_______UUCUGCAAAUAAAAUUUUGUUGCAUAUUUUGCGGCGUCAAAUGGCAGCCACAAAAAUGAAUGAAAUCGUAAAAUUCAGACA ....................((..((((((((.(((.(((((.((((((....))).))).))))))))))))))))..))........... (-15.19 = -15.33 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:59 2006