
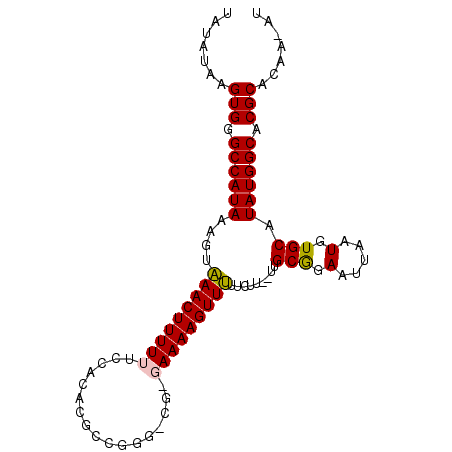
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,386,676 – 7,386,769 |
| Length | 93 |
| Max. P | 0.806991 |

| Location | 7,386,676 – 7,386,769 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -18.01 |
| Energy contribution | -17.76 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.806991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7386676 93 + 23771897 -AAAUAAGUGGGCCAUAAAGUGAACUUUUUUCCACACGCCGUCCCGG-AAAAGUUCUUCUU--GUGCGGAAUUAAUGUGCAUAUGGCACGCACAA-AU -......(((.(((((((((.((((((((((..((.....))...))-))))))))..)))--(..((.......))..).)))))).)))....-.. ( -24.40) >DroPse_CAF1 31540 93 + 1 UGUGUAAGUGGGCCAUAAAGUAAACUUUUUUCCACGCGCUGGG-UG-GAAAAGUUUUUCAU--UUGCGGAAUUAAUGUGCAUAUGGCACGCACAA-AU (((((..(((((....((((....))))..))))).(((.(((-((-(((.....))))))--)))))........((((.....))))))))).-.. ( -25.20) >DroGri_CAF1 31501 87 + 1 CUUGUAAGUGGGCCAUAUAACAAACUUUUUUACACAC-----------AAAAGUUUUGUUGGCCCGCAGAAUUAAUGUGCAUAUGGCACGCACCAAAU ..(((..((((((((....((((((((((........-----------))))).))))))))))))).........((((.....)))))))...... ( -26.30) >DroEre_CAF1 34884 93 + 1 -AAAUAAGUGGGCCAUAAAGUGAACUUUUUCCCACACGCCGUGCCG-GAAAAGUUCUCCUU--GCGCGGAAUUAAUGUGCAUAUGGCACGCACAA-AU -......(((((....((((....))))..)))))..((.((((((-((.......))).(--(((((.......))))))...)))))))....-.. ( -31.50) >DroAna_CAF1 41387 93 + 1 UAUAUAAGUGGGCCAUAAACUAAACUUUUUUCCACACGCCGA-CCG-GAAAAGUUUUCCUU--UUGCGGAAUUAAUGUGCAUAUGGCACGCACAA-AU .......(((.((((((...............((((......-(((-.(((((.....)))--)).)))......))))..)))))).)))....-.. ( -19.73) >DroPer_CAF1 31862 93 + 1 UGUGUAAGUGGGCCAUAAAGUAAACUUUUUUCCACGCGCUGGG-UG-GAAAAGUUUUUCAU--UUGCGGAAUUAAUGUGCAUAUGGCACGCACAA-AU (((((..(((((....((((....))))..))))).(((.(((-((-(((.....))))))--)))))........((((.....))))))))).-.. ( -25.20) >consensus UAUAUAAGUGGGCCAUAAAGUAAACUUUUUUCCACACGCCGGG_CG_GAAAAGUUUUUCUU__UUGCGGAAUUAAUGUGCAUAUGGCACGCACAA_AU .......(((.((((((....(((((((((.................))))))))).........(((.(.....).))).)))))).)))....... (-18.01 = -17.76 + -0.25)

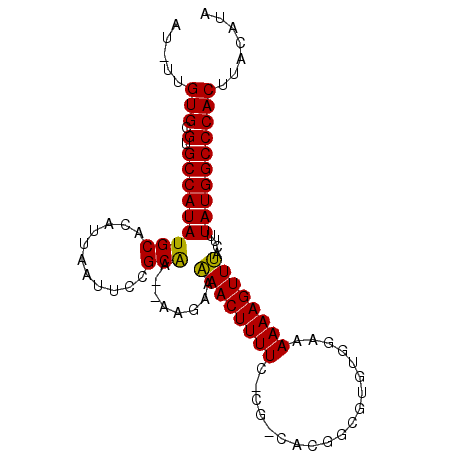
| Location | 7,386,676 – 7,386,769 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -16.01 |
| Energy contribution | -15.34 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7386676 93 - 23771897 AU-UUGUGCGUGCCAUAUGCACAUUAAUUCCGCAC--AAGAAGAACUUUU-CCGGGACGGCGUGUGGAAAAAAGUUCACUUUAUGGCCCACUUAUUU- ..-..(((.(.(((((...........((((((((--(((.....)))..-(((...))).)))))))).((((....)))))))))))))......- ( -22.60) >DroPse_CAF1 31540 93 - 1 AU-UUGUGCGUGCCAUAUGCACAUUAAUUCCGCAA--AUGAAAAACUUUUC-CA-CCCAGCGCGUGGAAAAAAGUUUACUUUAUGGCCCACUUACACA ..-..(((.(.(((((((((...........))).--.....(((((((((-((-(.......)))))..)))))))....))))))))))....... ( -22.00) >DroGri_CAF1 31501 87 - 1 AUUUGGUGCGUGCCAUAUGCACAUUAAUUCUGCGGGCCAACAAAACUUUU-----------GUGUGUAAAAAAGUUUGUUAUAUGGCCCACUUACAAG ..((((((((((...)))))))...........(((((((((((.(((((-----------........))))))))))....)))))).....))). ( -25.60) >DroEre_CAF1 34884 93 - 1 AU-UUGUGCGUGCCAUAUGCACAUUAAUUCCGCGC--AAGGAGAACUUUUC-CGGCACGGCGUGUGGGAAAAAGUUCACUUUAUGGCCCACUUAUUU- ..-..(((.(.((((((...........(((....--..)))(((((((((-(.((((...)))).)).))))))))....))))))))))......- ( -30.60) >DroAna_CAF1 41387 93 - 1 AU-UUGUGCGUGCCAUAUGCACAUUAAUUCCGCAA--AAGGAAAACUUUUC-CGG-UCGGCGUGUGGAAAAAAGUUUAGUUUAUGGCCCACUUAUAUA ..-..(((.(.((((((.((........(((....--..)))(((((((((-((.-........))))..))))))).)).))))))))))....... ( -22.40) >DroPer_CAF1 31862 93 - 1 AU-UUGUGCGUGCCAUAUGCACAUUAAUUCCGCAA--AUGAAAAACUUUUC-CA-CCCAGCGCGUGGAAAAAAGUUUACUUUAUGGCCCACUUACACA ..-..(((.(.(((((((((...........))).--.....(((((((((-((-(.......)))))..)))))))....))))))))))....... ( -22.00) >consensus AU_UUGUGCGUGCCAUAUGCACAUUAAUUCCGCAA__AAGAAAAACUUUUC_CG_CACGGCGUGUGGAAAAAAGUUUACUUUAUGGCCCACUUACAUA .....(((.(.(((((((((...........)))........((((((((...................))))))))....))))))))))....... (-16.01 = -15.34 + -0.66)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:44 2006