
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 753,085 – 753,185 |
| Length | 100 |
| Max. P | 0.851559 |

| Location | 753,085 – 753,185 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 75.06 |
| Mean single sequence MFE | -16.23 |
| Consensus MFE | -12.10 |
| Energy contribution | -12.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 753085 100 - 23771897 AAAAAA--CUUAUUAA-------AAAAAAACUGGACCAAAAAAUAUAUAGUCACCAACGACUUGCUCCAGGUGAGGCUCGAACUCACAACCCCGGCAUAGCUCAUACAC ......--........-------.......(((((.((..........((((......)))))).)))))(((((.......)))))...................... ( -16.60) >DroVir_CAF1 128195 95 - 1 AAUAAAAAUA----AAGCG---UAAAAAAA------UAAA-AAAGAACAGUCGACAACGACCUGCUCCAGGUGAGGCUCGAACUCACAACCCCGGCAUUGCUCAUACAC ..........----.((((---........------....-........((((....)))).((((....(((((.......)))))......)))).))))....... ( -16.00) >DroPse_CAF1 118194 100 - 1 AAAGAA---------AGAAAAUGAAGAAAAGGAGGGCAAAAAAAAUUAAGUCUCAACCGACCUGCUCCAGGUGAGGCUCGAACUCACAACCCCGGCAUUGCUCAUACAC ......---------..................((((((..........(((......))).((((....(((((.......)))))......))))))))))...... ( -19.50) >DroGri_CAF1 144332 95 - 1 AUGGAAAAUU----AAUUG---AAAAAAAA------UAUG-AAAAAAUUGUCGACAAAGACCUGCUCCAGGUGAGGCUCGAACUCACAACCCCGGCAUAGCUCAUACAC ..........----.....---........------((((-(......(((((......(((((...)))))(((.......))).......)))))....)))))... ( -15.60) >DroSec_CAF1 108214 108 - 1 UAUAAAGUCUUAUUAAGAA-AAAAAAAAAACUGUAUCAAAAAAUACAUAGUCACCAGCGACUUGCUCCAGGUGAGGCUCGAACUCACAACCCCGGCAUAGCUCAUACAC ....(((((..........-...........(((((......)))))..((.....)))))))(((....(((((.......)))))......)))............. ( -15.10) >DroEre_CAF1 113778 108 - 1 AUACAAAACUAUUUAAGAAAAAAAAAAAAACUGCACCAAA-AAGAUAUAGUCACCAGCGACUUGCUCCAGGUGAGGCUCGAACUCACAACCCCGGCAUAGCUCAUACAC ................................((......-.......((((......))))((((....(((((.......)))))......))))..))........ ( -14.60) >consensus AAAAAAAACU____AAGAA___AAAAAAAACUG_A_CAAA_AAAAUAUAGUCACCAACGACCUGCUCCAGGUGAGGCUCGAACUCACAACCCCGGCAUAGCUCAUACAC .................................................(((......))).((((....(((((.......)))))......))))............ (-12.10 = -12.10 + -0.00)

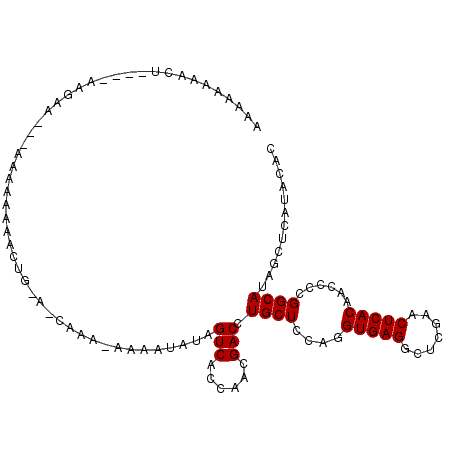
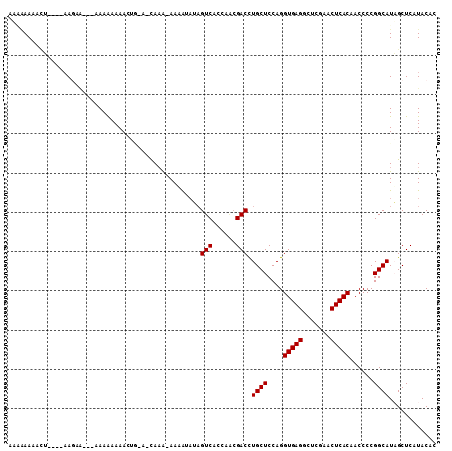
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:50 2006