
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,350,258 – 7,350,375 |
| Length | 117 |
| Max. P | 0.944746 |

| Location | 7,350,258 – 7,350,375 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.63 |
| Mean single sequence MFE | -22.06 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.30 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7350258 117 + 23771897 CUUUCAGCAGCUUUAUCUGCC--AAAAUGUAGACU-UAUCUAUUGAAAUGGCCAUGACAAUAGAAAAACAAGCAUAGUCUGUAAUGAUAAAUGCAUUAUCGAAAUAAAAAGCCACUGAUA ...((((..(((((.(((((.--.....)))))..-..(((((((..((....))..))))))).......((((.(((......)))..)))).............)))))..)))).. ( -23.50) >DroSec_CAF1 1123 118 + 1 UAUUCAGCAGCUUUAUCUGCC--AAAAUGUAGGCUUUAUCUAUUGAAAUGGCCAUGACAAUAGAAAAACAAGCAUAGUCUGUAAUGAUAAAUGCAUUAUCGAAAUAAAAAGCCACUGAUA ...(((((((......)))).--........((((((.(((((((..((....))..))))))).......((((.(((......)))..)))).............))))))..))).. ( -23.80) >DroSim_CAF1 1116 117 + 1 UAUUCAGCAGCUUUAUCUGCC--AAAAUGUAGGCU-UAUCUAUUGAAAUGGCCAUGACAAAAGAAAAACAAGCAUAGUCUGUAAUGAUAAAUGCAUUAUCGAAAUAAAAAGCCACUGAUA ...((((..(((((.......--....(((.((((-..((....))...))))...)))............((((.(((......)))..)))).............)))))..)))).. ( -21.30) >DroEre_CAF1 2183 116 + 1 CUUUUAGCGGCUUUAUCUGCC--AAAAUGUAGACU-UAUCUAUUGAACUGGCCAUGACAAAAGAA-AACAAGCACAGCCUGUAAUGAUAAAUGCAUUAUCGAAAUAAAAAGCGACUGAUA ......(((((.......)))--..((((((...(-((((.((((....(((..((.(.......-.....)))..)))..))))))))).)))))).............))........ ( -19.10) >DroYak_CAF1 1128 118 + 1 CUUUCAGUAGCUUUAUCUGCCCAAAAAUAUAGACU-UAUCUAUUGAAGUGGCCAUGACAAAAGAA-AACAAACCCAGUCUGUAAUGAUAAAUGCAUUAUCGAAAUAAAAAGCCACUGAUA ...(((((.(((((.((((.(((.....(((((..-..))))).....))).)).))........-...........((.((((((.......)))))).)).....))))).))))).. ( -22.60) >consensus CUUUCAGCAGCUUUAUCUGCC__AAAAUGUAGACU_UAUCUAUUGAAAUGGCCAUGACAAAAGAAAAACAAGCAUAGUCUGUAAUGAUAAAUGCAUUAUCGAAAUAAAAAGCCACUGAUA ...((((..(((((....(((.......(((((.....)))))......))).........................((.((((((.......)))))).)).....)))))..)))).. (-17.66 = -17.30 + -0.36)



| Location | 7,350,258 – 7,350,375 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.63 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -21.76 |
| Energy contribution | -21.64 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7350258 117 - 23771897 UAUCAGUGGCUUUUUAUUUCGAUAAUGCAUUUAUCAUUACAGACUAUGCUUGUUUUUCUAUUGUCAUGGCCAUUUCAAUAGAUA-AGUCUACAUUUU--GGCAGAUAAAGCUGCUGAAAG ..((((..(((.........(((((.((((...((......))..)))))))))......(((((...((((......((((..-..)))).....)--))).))))))))..))))... ( -27.80) >DroSec_CAF1 1123 118 - 1 UAUCAGUGGCUUUUUAUUUCGAUAAUGCAUUUAUCAUUACAGACUAUGCUUGUUUUUCUAUUGUCAUGGCCAUUUCAAUAGAUAAAGCCUACAUUUU--GGCAGAUAAAGCUGCUGAAUA ..((((..(((((.....(((((((.((((...((......))..)))))))))..(((((((..((....))..)))))))....(((........--))).)).)))))..))))... ( -29.90) >DroSim_CAF1 1116 117 - 1 UAUCAGUGGCUUUUUAUUUCGAUAAUGCAUUUAUCAUUACAGACUAUGCUUGUUUUUCUUUUGUCAUGGCCAUUUCAAUAGAUA-AGCCUACAUUUU--GGCAGAUAAAGCUGCUGAAUA ..((((..(((((.......(((((.((((...((......))..))))))))).....(((((((.(((....((....))..-.))).......)--)))))).)))))..))))... ( -27.10) >DroEre_CAF1 2183 116 - 1 UAUCAGUCGCUUUUUAUUUCGAUAAUGCAUUUAUCAUUACAGGCUGUGCUUGUU-UUCUUUUGUCAUGGCCAGUUCAAUAGAUA-AGUCUACAUUUU--GGCAGAUAAAGCCGCUAAAAG ....(((.((..........(((((.....)))))...((((((...)))))).-....((((((...(((((.....((((..-..))))....))--))).)))))))).)))..... ( -23.60) >DroYak_CAF1 1128 118 - 1 UAUCAGUGGCUUUUUAUUUCGAUAAUGCAUUUAUCAUUACAGACUGGGUUUGUU-UUCUUUUGUCAUGGCCACUUCAAUAGAUA-AGUCUAUAUUUUUGGGCAGAUAAAGCUACUGAAAG ..((((((((..........(((((.....)))))...((((((...)))))).-....((((((.((.(((.....(((((..-..))))).....))).))))))))))))))))... ( -31.00) >consensus UAUCAGUGGCUUUUUAUUUCGAUAAUGCAUUUAUCAUUACAGACUAUGCUUGUUUUUCUUUUGUCAUGGCCAUUUCAAUAGAUA_AGUCUACAUUUU__GGCAGAUAAAGCUGCUGAAAG ..(((((((((.........(((((.((((...((......))..)))))))))......(((((...(((.......(((.......)))........))).))))))))))))))... (-21.76 = -21.64 + -0.12)

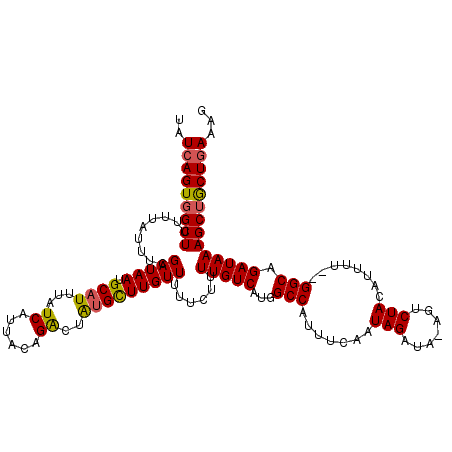

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:33 2006