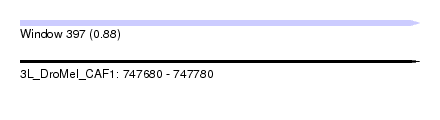
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 747,680 – 747,780 |
| Length | 100 |
| Max. P | 0.883439 |

| Location | 747,680 – 747,780 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
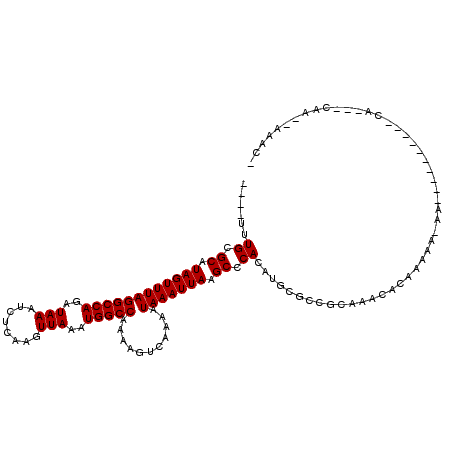
| Mean single sequence MFE | -17.08 |
| Consensus MFE | -12.48 |
| Energy contribution | -12.48 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 747680 100 + 23771897 ----UUUGCGCAUAGUUUAGGCCAGAUAAAUCUUAAGUUAAAUGGCCAAAAGUCAAAAUAAAUUAAGCCCACAUGCGCCGAAAACACAACAAAAA----------CA---CAA--AAGU- ----...((((((.((((((((((..(((........)))..)))))....((....))....)))))....)))))).................----------..---...--....- ( -16.30) >DroPse_CAF1 112727 101 + 1 ----UUUGCGCAUAGUUUAGGCCAGAUAAAUCUCGAGUUAAAUGGCCAAAAGUCAAAAUAAAUUAAGCCCACAUGCGCCGCAAACACAAACA-AA--------CACA---CAC--AGAU- ----(((((((........(((((..(((........)))..)))))...................((......))).))))))........-..--------....---...--....- ( -16.80) >DroGri_CAF1 139610 98 + 1 UGCCUGUGCGCAUAGUUUAGGCCAGAUAAAUCUCAAGUUAAAUGGCCAAAAGUCAAAAUAAAUUAAGCCCACAUGCAGCCAAACUAAAAACA-A---------------------AAAAA (((.((((.((.((((((((((((..(((........)))..)))))...........))))))).)).)))).)))...............-.---------------------..... ( -21.30) >DroWil_CAF1 103094 101 + 1 UCUCUCUGCGCAUAGUUUAGGCCAGAUAAAUCUCAAGUUAAAUGGCCAAAAGUCAAAAUAAAUUAAGCCCACAUGCGGU----ACACAAGAA-AA----------UC---AAACCAAGC- .....((((((.((((((((((((..(((........)))..)))))...........))))))).))......)))).----.........-..----------..---.........- ( -13.90) >DroAna_CAF1 110811 106 + 1 ----UUUGCGCAUAGUUUAGGCCAGAUAAAUCUCAAGUUAAAUGGCCAAAAGUCAAAAUAAAUUAAGCCCACAUGCGACACAAAAAUAACAA-CAG------AAACGGGCCAG--AAAC- ----((((.((...((((.(((((..(((........)))..)))))....(((((......))..((......))))).............-...------))))..)))))--)...- ( -17.40) >DroPer_CAF1 118311 109 + 1 ----UUUGCGCAUAGUUUAGGCCAGAUAAAUCUCGAGUUAAAUGGCCAAAAGUCAAAAUAAAUUAAGCCCACAUGCGCCGCAAACACAAACA-AACACACAAACACA---CAC--AGAC- ----(((((((........(((((..(((........)))..)))))...................((......))).))))))........-..............---...--....- ( -16.80) >consensus ____UUUGCGCAUAGUUUAGGCCAGAUAAAUCUCAAGUUAAAUGGCCAAAAGUCAAAAUAAAUUAAGCCCACAUGCGCCGCAAACACAAAAA_AA__________CA___CAA__AAAC_ ......((.((.((((((((((((..(((........)))..)))))...........))))))).)).))................................................. (-12.48 = -12.48 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:49 2006