| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,340,501 – 7,340,621 |
| Length | 120 |
| Max. P | 0.999874 |

| Location | 7,340,501 – 7,340,621 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
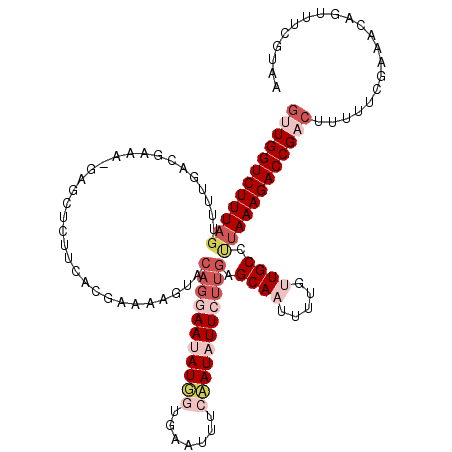
| Reading direction | forward |
| Mean pairwise identity | 75.67 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -16.06 |
| Energy contribution | -16.62 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7340501 120 + 23771897 UUACGAAACUGUUUCGAAAAAGUCGGUCUUUAAGGCAACAAAAUUGCUCAAGAAUAUUGAAAUUCACCAUAUUCCUGUACUUUUCGUGAAAAGUUUUUUUCGUCAAACCUAAAGACCAAC ...((((.....))))........((((((((.(((((.....)))))((.((((((((.....))..)))))).)).((((((....))))))...............))))))))... ( -21.10) >DroSec_CAF1 5500 119 + 1 UUACGAAACUGUUUCGAAAAAGUCGGUCUUUAAGGCAAAGAAAUUGCUCAAGAAUAUGGAAAUUCACCAUAUUCCUGUACUUUUCGUGAAGAGCUC-UUUCGUCAAAACUAAAGACCAAC ...((((.....))))........((((((((.......((((..((((..((((((((.......))))))))...(((.....)))..))))..-))))........))))))))... ( -30.46) >DroSim_CAF1 5561 119 + 1 UUACGAAACUGUUUCGAAAUAGUCGGUCUUUAAGGCAACGAAAUUGCUCAAGAAUAUGGAAAUUCACCAUAUUCCUGUACUUCUCGCGAAGAGCUC-UUUCGUCAAAACUAAAGACCAAC .......((((((....)))))).((((((((.....((((((..((((..((((((((.......))))))))......(((....)))))))..-))))))......))))))))... ( -33.80) >DroEre_CAF1 5628 111 + 1 UUACGAAACUGCUUCGAAAAAGCCGGUCUUUAGGGCAACAAAAGUGCUCAAGAAUAUUUCAGUUCAUAAUCUUUGUGCGCUAUUUGUAAAGAGCUA-CC------UAACUAAAGACCA-- ...((((.....))))........(((((((((....((((((((((.(((((.(((........))).)).))).))))).)))))..((.....-.)------)..))))))))).-- ( -27.60) >DroYak_CAF1 5599 93 + 1 UUACGAAACU--------AAAGUCGGUCUUUAGAGCAACAAACGUGCUCAAGAAUAUUGCAGUUCAUAAUCUUUC--------UUGCAAAGAGCUA-CC------U----AAAGACCAGC ..........--------......((((((((((((.......(.((((((.....))).))).)....(((((.--------....)))))))).-.)------)----)))))))... ( -20.70) >consensus UUACGAAACUGUUUCGAAAAAGUCGGUCUUUAAGGCAACAAAAUUGCUCAAGAAUAUUGAAAUUCACCAUAUUCCUGUACUUUUCGUGAAGAGCUA_UUUCGUCAAAACUAAAGACCAAC ........................((((((((.(((((.....)))))...((((((((.......))))))))...................................))))))))... (-16.06 = -16.62 + 0.56)

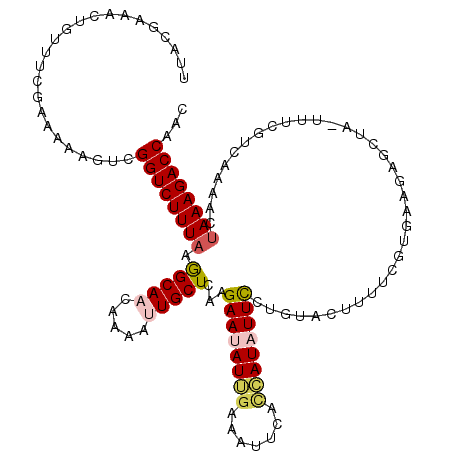
| Location | 7,340,501 – 7,340,621 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.67 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -19.46 |
| Energy contribution | -22.42 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.57 |
| SVM decision value | 4.34 |
| SVM RNA-class probability | 0.999874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7340501 120 - 23771897 GUUGGUCUUUAGGUUUGACGAAAAAAACUUUUCACGAAAAGUACAGGAAUAUGGUGAAUUUCAAUAUUCUUGAGCAAUUUUGUUGCCUUAAAGACCGACUUUUUCGAAACAGUUUCGUAA ((((((((((((((..(((((((...((((((....)))))).(((((((((((......)).))))))))).....)))))))))).))))))))))).....((((.....))))... ( -36.40) >DroSec_CAF1 5500 119 - 1 GUUGGUCUUUAGUUUUGACGAAA-GAGCUCUUCACGAAAAGUACAGGAAUAUGGUGAAUUUCCAUAUUCUUGAGCAAUUUCUUUGCCUUAAAGACCGACUUUUUCGAAACAGUUUCGUAA ((((((((((((....((.((((-..((((...((.....))..((((((((((.......))))))))))))))..))))))....)))))))))))).....((((.....))))... ( -35.70) >DroSim_CAF1 5561 119 - 1 GUUGGUCUUUAGUUUUGACGAAA-GAGCUCUUCGCGAGAAGUACAGGAAUAUGGUGAAUUUCCAUAUUCUUGAGCAAUUUCGUUGCCUUAAAGACCGACUAUUUCGAAACAGUUUCGUAA ((((((((((((...((((((((-..(((((((....))))..(((((((((((.......))))))))))))))..))))))))..)))))))))))).....((((.....))))... ( -46.50) >DroEre_CAF1 5628 111 - 1 --UGGUCUUUAGUUA------GG-UAGCUCUUUACAAAUAGCGCACAAAGAUUAUGAACUGAAAUAUUCUUGAGCACUUUUGUUGCCCUAAAGACCGGCUUUUUCGAAGCAGUUUCGUAA --.(((((((((...------((-((((...........((.((...((((.(((........))).))))..)).))...))))))))))))))).(((((...))))).......... ( -28.04) >DroYak_CAF1 5599 93 - 1 GCUGGUCUUU----A------GG-UAGCUCUUUGCAA--------GAAAGAUUAUGAACUGCAAUAUUCUUGAGCACGUUUGUUGCUCUAAAGACCGACUUU--------AGUUUCGUAA ..((((((((----(------((-((((....(((..--------..((((.(((........))).))))..))).....))))).)))))))))).....--------.......... ( -25.50) >consensus GUUGGUCUUUAGUUUUGACGAAA_GAGCUCUUCACGAAAAGUACAGGAAUAUGGUGAAUUUCAAUAUUCUUGAGCAAUUUUGUUGCCUUAAAGACCGACUUUUUCGAAACAGUUUCGUAA ((((((((((((...............................(((((((((((.......))))))))))).((((.....)))).))))))))))))..................... (-19.46 = -22.42 + 2.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:28 2006