
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,294,111 – 7,294,209 |
| Length | 98 |
| Max. P | 0.523970 |

| Location | 7,294,111 – 7,294,209 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.38 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -17.58 |
| Energy contribution | -19.47 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7294111 98 + 23771897 AACAUGCAAAAUAAUCGGCAUGAUACACGUGGAUGCGCUGCCAGGUGAGAGAAGCUACACCAAAGCCUCAGAAAC-CAACAAA-------UCCGAUUGCUACUAAU ...........(((((((..((...(((.(((........))).))).(((..(((.......))))))......-...))..-------.)))))))........ ( -19.20) >DroVir_CAF1 131480 99 + 1 GAAAUGCAAAAUUAUUGGCAUGAUACAUGUGGAUGCGCUGCCAGGUAAGGC----CGUGCCGCA--UGCAGCAGC-AAACAAACGCAUUUUCGCAUUGAUUUUAAU (((((((......(((.(((((...))))).)))(((((((..((((....----..))))...--.))))).))-........)))))))............... ( -26.50) >DroSec_CAF1 52121 98 + 1 AACAUGCAAAGUAAUCGGCAUGAUACACGUGGAUGCGCUGCCAGGUGAGAGAGGCUAGACCAGAGCCUCAGUAAC-CACCAAA-------UCCGAUUGCUACUAAU .........(((((((((..((...(((.(((........))).)))...((((((.......))))))......-...))..-------.)))))))))...... ( -30.00) >DroSim_CAF1 55663 98 + 1 AACAUGCAAAGUAAUCGGCAUGAUACACGUGGAUGCGCUGCCAGGUGAGAGAGUCUACACCAGAGCCUCAGUAAC-CACCAAA-------UCCGAUUGCUACUAAU .........(((((((((.((.......((((.(((...(((.((((((.....)).)))).).))....))).)-)))...)-------))))))))))...... ( -26.80) >DroEre_CAF1 52444 98 + 1 AACAUGCAAAGUAAUCGGCAUGAUACACGUGGAUGCGCUGCCAGGUGAGAGAAGCUACACCAGAGCUUCAGAACC-AACGAAA-------UCCGAUUGCUUCUAAU ........((((((((((.......(((.(((........))).)))...((((((.......))))))......-.......-------.))))))))))..... ( -27.90) >DroYak_CAF1 53304 99 + 1 AACAUGCAAAGUAAUCGGCAUGAUACACGUGGAUGCGCUGCCAGGUGAGAGAAGCUACGCCAGAGCUACAGAACCGAACGAAA-------UCCGAUUGCUUUUAAU .......(((((((((((.......(((.(((........))).)))...(.((((.(....))))).)..............-------.))))))))))).... ( -24.90) >consensus AACAUGCAAAGUAAUCGGCAUGAUACACGUGGAUGCGCUGCCAGGUGAGAGAAGCUACACCAGAGCCUCAGAAAC_AACCAAA_______UCCGAUUGCUACUAAU .........(((((((((.......(((.(((........))).)))...((((((.......))))))......................)))))))))...... (-17.58 = -19.47 + 1.89)

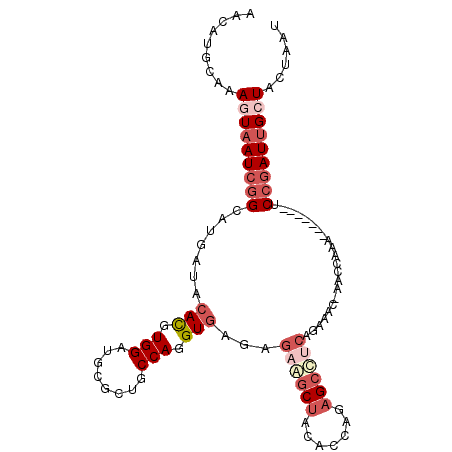
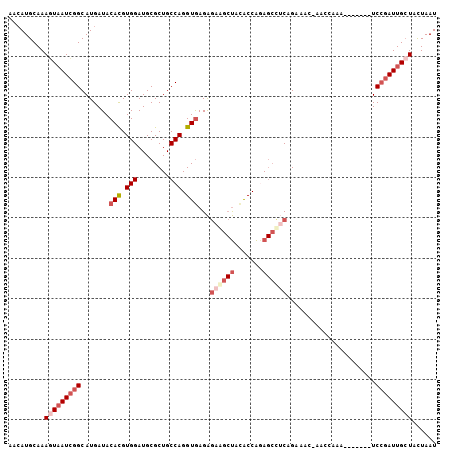
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:20 2006