
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
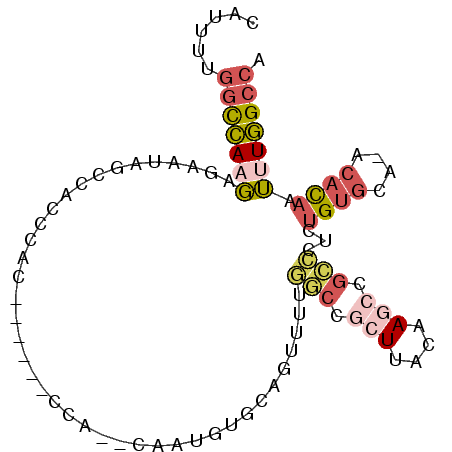
| Location | 7,291,159 – 7,291,253 |
| Length | 94 |
| Max. P | 0.934588 |

| Location | 7,291,159 – 7,291,253 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 57.05 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -9.38 |
| Energy contribution | -10.74 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7291159 94 + 23771897 CAUUUUGGCCAAGAGAAUAGCCACCUACUGUGCUCCA-UCGAUUUGCAGUUUUGGCCACUUACAAGCCGCCCUCUGUGCACACACAUUUUGGCCA .....(((((((((.....((((...(((((..((..-..))...)))))..))))..................((((....))))))))))))) ( -24.50) >DroSim_CAF1 52750 77 + 1 UAUAUUGGCCAAGAGGAUAG--ACCCAC------CCA-ACGCUUUGCAGUUUUGGCCGCUUACAA---------UGUGCACACACAUUUUGGCCA ......((((((((((....--..))..------...-..((...))..))))))))(((...((---------((((....))))))..))).. ( -21.00) >DroEre_CAF1 49497 79 + 1 CAUUUUGGCCAAGGGGACAGCCACCC--------------AAUGUGCAGUUUUGGCCGCUUACAAGCCGCCCUCUGUGC--ACACAAUGUGGCCA .....((((((.(((........)))--------------..((..(((....(((.(((....))).)))..)))..)--).......)))))) ( -30.80) >DroYak_CAF1 50354 93 + 1 CAAUUUGGCCAAGAACAUAGCCACCCACUGUGGGCCAUUCAAUGUGCAGUUUUGGCCGCUUACAAGCCGCCCUCUGUGC--ACACAAUUUGGCCA .....((((((((.......((((.....)))).........((..(((....(((.(((....))).)))..)))..)--).....)))))))) ( -31.40) >DroMoj_CAF1 94825 77 + 1 UUUGCAGCUAAUCAAAUUGCAUGUGU--------------AUUCGGUCUUUCGUCCAGGUAUUCAGCUGGG--CUGACUAGGCAU--GUUUGCAA .((((((...........((((((.(--------------(.((((....))((((((........)))))--).)).)).))))--)))))))) ( -20.40) >consensus CAUUUUGGCCAAGAGAAUAGCCACCCAC______CCA__CAAUGUGCAGUUUUGGCCGCUUACAAGCCGCCCUCUGUGCA_ACACAAUUUGGCCA ......(((((((........................................(((.(((....))).)))...((((....)))).))))))). ( -9.38 = -10.74 + 1.36)

| Location | 7,291,159 – 7,291,253 |
|---|---|
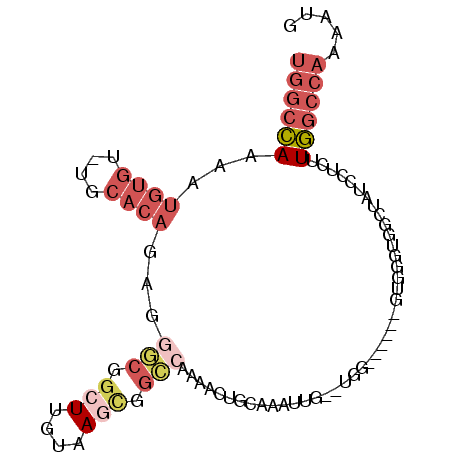
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 57.05 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -10.32 |
| Energy contribution | -12.64 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.36 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7291159 94 - 23771897 UGGCCAAAAUGUGUGUGCACAGAGGGCGGCUUGUAAGUGGCCAAAACUGCAAAUCGA-UGGAGCACAGUAGGUGGCUAUUCUCUUGGCCAAAAUG ..(((....((((....))))...)))((((.(..((((((((..((((....((..-..))...))))...))))))))..)..))))...... ( -32.10) >DroSim_CAF1 52750 77 - 1 UGGCCAAAAUGUGUGUGCACA---------UUGUAAGCGGCCAAAACUGCAAAGCGU-UGG------GUGGGU--CUAUCCUCUUGGCCAAUAUA (((((((((((((....))))---------))....((((......)))).......-.((------(((...--.)))))..)))))))..... ( -24.70) >DroEre_CAF1 49497 79 - 1 UGGCCACAUUGUGU--GCACAGAGGGCGGCUUGUAAGCGGCCAAAACUGCACAUU--------------GGGUGGCUGUCCCCUUGGCCAAAAUG ((((((....((((--(((.....(((.(((....))).))).....))))))).--------------(((........))).))))))..... ( -32.60) >DroYak_CAF1 50354 93 - 1 UGGCCAAAUUGUGU--GCACAGAGGGCGGCUUGUAAGCGGCCAAAACUGCACAUUGAAUGGCCCACAGUGGGUGGCUAUGUUCUUGGCCAAAUUG .(((((....((((--(((.....(((.(((....))).))).....)))))))....)))))..((((...((((((......)))))).)))) ( -39.70) >DroMoj_CAF1 94825 77 - 1 UUGCAAAC--AUGCCUAGUCAG--CCCAGCUGAAUACCUGGACGAAAGACCGAAU--------------ACACAUGCAAUUUGAUUAGCUGCAAA (((((...--..((.(((((((--....((((..((..(((.(....).)))..)--------------)..)).))...)))))))))))))). ( -15.30) >consensus UGGCCAAAAUGUGU_UGCACAGAGGGCGGCUUGUAAGCGGCCAAAACUGCAAAUUG__UGG______GUGGGUGGCUAUCCUCUUGGCCAAAAUG ((((((...((((....))))...(((.(((....))).)))..........................................))))))..... (-10.32 = -12.64 + 2.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:19 2006