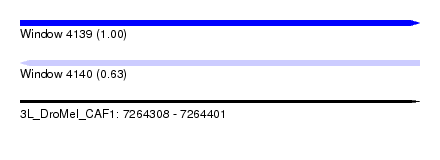
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,264,308 – 7,264,401 |
| Length | 93 |
| Max. P | 0.999974 |

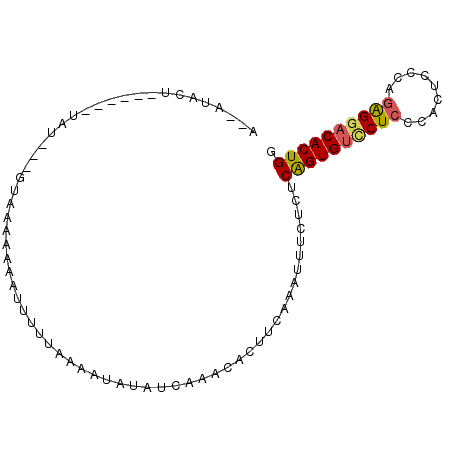
| Location | 7,264,308 – 7,264,401 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 69.71 |
| Mean single sequence MFE | -17.20 |
| Consensus MFE | -12.59 |
| Energy contribution | -12.77 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.73 |
| SVM decision value | 5.11 |
| SVM RNA-class probability | 0.999974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7264308 93 + 23771897 UUACUACUGUAAGGUAUACUGUAAAAGAAUUUUUAAAAUAUAUCAAACACUUUAAAUUUCUCUCAGUGUCCUCCCACUCCCAGAGGACACUGG ((((....))))((((((...(((((....)))))...))))))...................((((((((((.........)))))))))). ( -22.40) >DroSim_CAF1 21238 81 + 1 U--CUACU------UAU----UCAAAAAAUAUUUAAAAUAUAUCAUAUACUUCAAAUUUCUCUCAGUGUUCUCCCACUCCCAGAGGACACUGG .--.....------...----..........................................((((((((((.........)))))))))). ( -14.00) >DroEre_CAF1 20919 82 + 1 A--AUACU------UGU---GAAAAAAAAUGUUUAUAACAGAGCAAUCACUUCAAAUUUCGCUCGGUGCACUCCCACUCCCAGGGGACACUGG .--.....------.((---((((.....(((.....)))((....))........)))))).(((((...((((........))))))))). ( -16.50) >DroYak_CAF1 21986 74 + 1 A--AUAUU------UA----GUAAAG-AAUUCUUUAAAUAUCCUAAUCGCUGCCACUUUCUCUCGGUGUCCUCCCAC------AGGACACUGG .--.....------..----.(((((-....)))))...........................(((((((((.....------))))))))). ( -15.90) >consensus A__AUACU______UAU___GUAAAAAAAUUUUUAAAAUAUAUCAAACACUUCAAAUUUCUCUCAGUGUCCUCCCACUCCCAGAGGACACUGG ...............................................................((((((((((.........)))))))))). (-12.59 = -12.77 + 0.19)

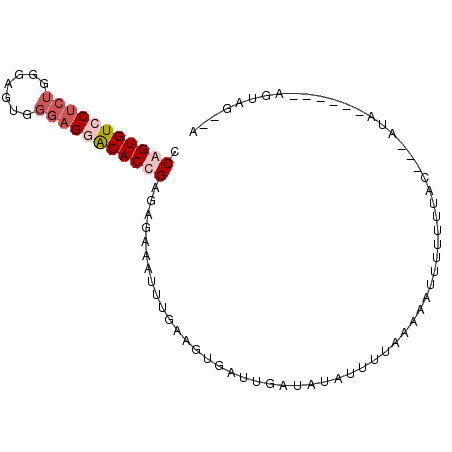
| Location | 7,264,308 – 7,264,401 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 69.71 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -10.04 |
| Energy contribution | -11.60 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7264308 93 - 23771897 CCAGUGUCCUCUGGGAGUGGGAGGACACUGAGAGAAAUUUAAAGUGUUUGAUAUAUUUUAAAAAUUCUUUUACAGUAUACCUUACAGUAGUAA .(((((((((((.......)))))))))))((((((.((((((((((.....))))))))))..))))))(((.(((.....))).))).... ( -28.60) >DroSim_CAF1 21238 81 - 1 CCAGUGUCCUCUGGGAGUGGGAGAACACUGAGAGAAAUUUGAAGUAUAUGAUAUAUUUUAAAUAUUUUUUGA----AUA------AGUAG--A .((((((.((((.......)))).))))))((((((((((((((((((...)))))))))))).))))))..----...------.....--. ( -20.00) >DroEre_CAF1 20919 82 - 1 CCAGUGUCCCCUGGGAGUGGGAGUGCACCGAGCGAAAUUUGAAGUGAUUGCUCUGUUAUAAACAUUUUUUUUC---ACA------AGUAU--U ((((......))))..(((..((......((((((.((.....))..))))))(((.....)))....))..)---)).------.....--. ( -15.60) >DroYak_CAF1 21986 74 - 1 CCAGUGUCCU------GUGGGAGGACACCGAGAGAAAGUGGCAGCGAUUAGGAUAUUUAAAGAAUU-CUUUAC----UA------AAUAU--U ((((((((((------.....)))))))(....)....)))..........(((((((((((....-)))...----))------)))))--) ( -15.20) >consensus CCAGUGUCCUCUGGGAGUGGGAGGACACCGAGAGAAAUUUGAAGUGAUUGAUAUAUUUUAAAAAUUUUUUUAC___AUA______AGUAG__A .(((((((((((.......)))))))))))............................................................... (-10.04 = -11.60 + 1.56)

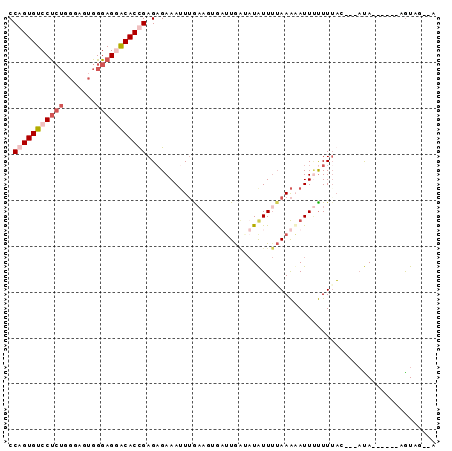
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:15 2006