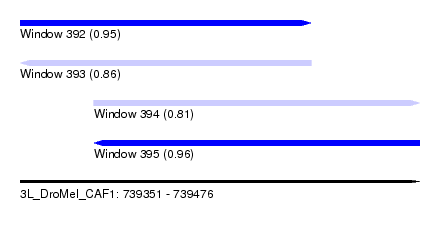
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 739,351 – 739,476 |
| Length | 125 |
| Max. P | 0.955544 |

| Location | 739,351 – 739,442 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.55 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -16.56 |
| Energy contribution | -16.53 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
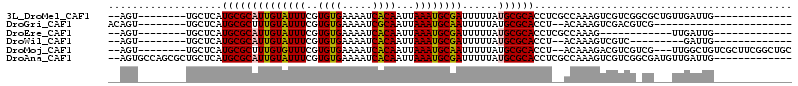
>3L_DroMel_CAF1 739351 91 + 23771897 -------------CAAUCAACAGCGCCGACGACUUUGGCGAGGUGCGCAUAAAAAUCGCAUUUAAUUGUGAUUUUCACACGAAAUACAAUGCGCAUGAGCA--------ACU-- -------------.........(((((((.....)))))...((((((((...(((((((......)))))))(((....))).....))))))))..)).--------...-- ( -26.70) >DroGri_CAF1 126180 81 + 1 -----------------------CGACGUCGACUUUGU--AGGUGCGCAUAAAAAUUGCAUUUAAUUGCGAUUUUCACACGAAAUACAAAGCGCAUGAGCA--------ACUGU -----------------------...(((((.((((((--(....((....(((((((((......)))))))))....))...))))))))).)))....--------..... ( -20.40) >DroEre_CAF1 99614 79 + 1 -------------CAAUCAA------------CUUUGGCGAGGUGCGCAUAAAAAUCGCAUUUAAUUGUGAUUUUCACACGAAAUACAAUGCGCAUGAGCA--------ACU-- -------------.......------------.....((...((((((((...(((((((......)))))))(((....))).....))))))))..)).--------...-- ( -20.70) >DroWil_CAF1 92817 80 + 1 -------------CAAUC---------GACGACUUUGU--AGGUGCGCAUAAAAAUCGCAUUUAAUUGUGAUUUUCACACGAAAUACAAUGCGCAUGAGCA--------ACU-- -------------.....---------.......((((--..((((((((...(((((((......)))))))(((....))).....))))))))..)))--------)..-- ( -21.10) >DroMoj_CAF1 109634 99 + 1 GCAGCCGAAGCGACAGCCAA---CGACGACGUCUUUGU--AGGUGCGCAUAAAAAUUGCAUUUAAUUGUGAUUUUCACACGAAACACAAAGCGCAUGAGCA--------ACU-- ((.......(((...(((.(---(((.(....).))))--.))).)))...(((((..((......))..)))))...............))((....)).--------...-- ( -20.60) >DroAna_CAF1 101853 99 + 1 -------------CAAUCAACAUCGCCGACGACUUUGGCGAGGUGCGCAUAAAAAUCGCAUUUAAUUGUGAUUUUCACACGAAAUACAAUGCGCAUGAGCAGCGCUGGCACU-- -------------.........(((((((.....)))))))(((((.((..(((((((((......)))))))))...............((((.......)))))))))))-- ( -30.00) >consensus _____________CAAUCAA___CG_CGACGACUUUGG__AGGUGCGCAUAAAAAUCGCAUUUAAUUGUGAUUUUCACACGAAAUACAAUGCGCAUGAGCA________ACU__ ..........................................((((((((...(((((((......)))))))(((....))).....)))))))).................. (-16.56 = -16.53 + -0.03)

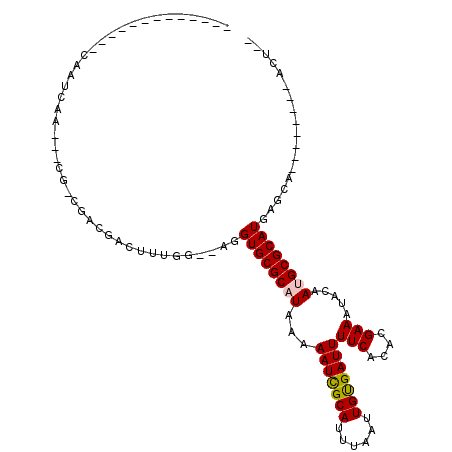
| Location | 739,351 – 739,442 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.55 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -17.03 |
| Energy contribution | -16.87 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 739351 91 - 23771897 --AGU--------UGCUCAUGCGCAUUGUAUUUCGUGUGAAAAUCACAAUUAAAUGCGAUUUUUAUGCGCACCUCGCCAAAGUCGUCGGCGCUGUUGAUUG------------- --...--------.((...((((((((((((((..((((.....))))...))))))))......))))))....(((.........))))).........------------- ( -23.00) >DroGri_CAF1 126180 81 - 1 ACAGU--------UGCUCAUGCGCUUUGUAUUUCGUGUGAAAAUCGCAAUUAAAUGCAAUUUUUAUGCGCACCU--ACAAAGUCGACGUCG----------------------- ...((--------(((....))((((((((...(((((((((((.(((......))).)))))))))))....)--))))))).)))....----------------------- ( -22.70) >DroEre_CAF1 99614 79 - 1 --AGU--------UGCUCAUGCGCAUUGUAUUUCGUGUGAAAAUCACAAUUAAAUGCGAUUUUUAUGCGCACCUCGCCAAAG------------UUGAUUG------------- --...--------.((...((((((((((((((..((((.....))))...))))))))......))))))....)).....------------.......------------- ( -18.20) >DroWil_CAF1 92817 80 - 1 --AGU--------UGCUCAUGCGCAUUGUAUUUCGUGUGAAAAUCACAAUUAAAUGCGAUUUUUAUGCGCACCU--ACAAAGUCGUC---------GAUUG------------- --(((--------((.....((((.(((((...((((((((((((.((......)).))))))))))))....)--)))).).))))---------)))).------------- ( -18.80) >DroMoj_CAF1 109634 99 - 1 --AGU--------UGCUCAUGCGCUUUGUGUUUCGUGUGAAAAUCACAAUUAAAUGCAAUUUUUAUGCGCACCU--ACAAAGACGUCGUCG---UUGGCUGUCGCUUCGGCUGC --((.--------(((.((((....((((((((..((((.....))))...))))))))....)))).))).))--.....(((((((...---.)))).)))((....))... ( -26.70) >DroAna_CAF1 101853 99 - 1 --AGUGCCAGCGCUGCUCAUGCGCAUUGUAUUUCGUGUGAAAAUCACAAUUAAAUGCGAUUUUUAUGCGCACCUCGCCAAAGUCGUCGGCGAUGUUGAUUG------------- --.((((((((((.......))))(((((((((..((((.....))))...))))))))).....)).)))).(((((.........))))).........------------- ( -30.70) >consensus __AGU________UGCUCAUGCGCAUUGUAUUUCGUGUGAAAAUCACAAUUAAAUGCGAUUUUUAUGCGCACCU__ACAAAGUCGUCG_CG___UUGAUUG_____________ ...................((((((((((((((..((((.....))))...))))))))......))))))........................................... (-17.03 = -16.87 + -0.16)
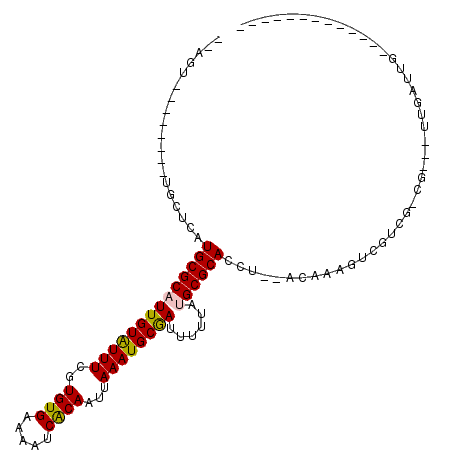
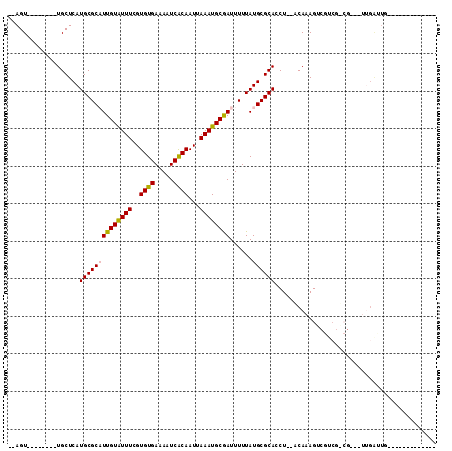
| Location | 739,374 – 739,476 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.34 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 739374 102 + 23771897 GGCGAG--GUGCGCAUAAAAAUCGCAUUUAAUUGUGAUUUUCACACGAAAUACAAUGCGCAUGAGCA--------ACUGGCGCCGC----UGGUUAAAAUGCUUAAAUCGAAGGGG ..(((.--((((((((...(((((((......)))))))(((....))).....))))))))(((((--------((..((...))----..)).....)))))...)))...... ( -28.60) >DroPse_CAF1 104440 93 + 1 GGCGAGGGGUGCGCAUAAAAAUCGCAUUUAAUUGUGAUUUUCACACGAAAUACAAUGCGCAUGAGCA--------ACUGGCGCCGC----UGCUUGACA----------GAGGG-G ((((..((((((((((...(((((((......)))))))(((....))).....)))))))).....--------.))..)))).(----(.(((....----------))).)-) ( -27.30) >DroWil_CAF1 92831 94 + 1 GU--AG--GUGCGCAUAAAAAUCGCAUUUAAUUGUGAUUUUCACACGAAAUACAAUGCGCAUGAGCA--------ACUGGCGCCCC----GGGG------GGUUAACCAGACAUGG ((--..--((((((((...(((((((......)))))))(((....))).....))))))))..)).--------.((((.((((.----...)------)))...))))...... ( -31.70) >DroMoj_CAF1 109667 92 + 1 GU--AG--GUGCGCAUAAAAAUUGCAUUUAAUUGUGAUUUUCACACGAAACACAAAGCGCAUGAGCA--------ACUGGCGCCAGUUGCUGGUUGCC------------GAGCCG ..--.(--(((((((.......)))......(((((..((((....))))))))).))((((.((((--------((((....)))))))).).))).------------..))). ( -30.20) >DroAna_CAF1 101876 99 + 1 GGCGAG--GUGCGCAUAAAAAUCGCAUUUAAUUGUGAUUUUCACACGAAAUACAAUGCGCAUGAGCAGCGCUGGCACUGGCGCCGC----UGCUUAGCA-----------GAGGCG .((.((--((((((((...(((((((......)))))))(((....))).....)))))))).....((....)).)).))(((.(----(((...)))-----------).))). ( -35.50) >DroPer_CAF1 109885 94 + 1 GGCGAGGGGUGCGCAUAAAAAUCGCAUUUAAUUGUGAUUUUCACACGAAAUACAAUGCGCAUGAGCA--------ACUGGCGCCGC----UGCUUGACA----------GAGGGGG ((((..((((((((((...(((((((......)))))))(((....))).....)))))))).....--------.))..)))).(----(.(((....----------))).)). ( -28.60) >consensus GGCGAG__GUGCGCAUAAAAAUCGCAUUUAAUUGUGAUUUUCACACGAAAUACAAUGCGCAUGAGCA________ACUGGCGCCGC____UGCUUGACA__________GAAGGGG (((.....((((((((...(((((((......)))))))(((....))).....))))))))..((.............)))))................................ (-18.64 = -19.34 + 0.70)

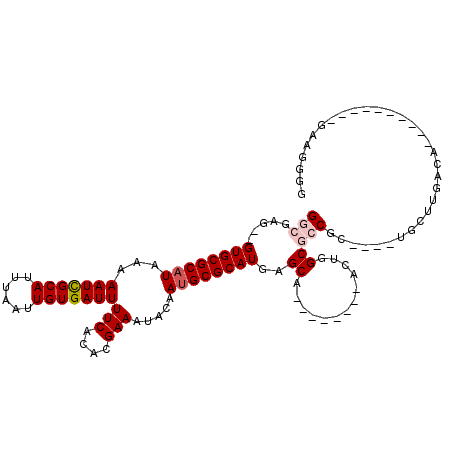
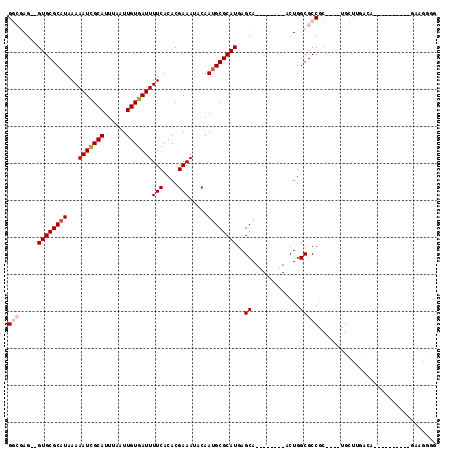
| Location | 739,374 – 739,476 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -18.74 |
| Energy contribution | -18.63 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 739374 102 - 23771897 CCCCUUCGAUUUAAGCAUUUUAACCA----GCGGCGCCAGU--------UGCUCAUGCGCAUUGUAUUUCGUGUGAAAAUCACAAUUAAAUGCGAUUUUUAUGCGCAC--CUCGCC ..............((.........(----(((((....))--------))))..((((((((((((((..((((.....))))...))))))))......)))))).--...)). ( -25.90) >DroPse_CAF1 104440 93 - 1 C-CCCUC----------UGUCAAGCA----GCGGCGCCAGU--------UGCUCAUGCGCAUUGUAUUUCGUGUGAAAAUCACAAUUAAAUGCGAUUUUUAUGCGCACCCCUCGCC .-.....----------......(((----(((((....))--------))))..((((((((((((((..((((.....))))...))))))))......))))))......)). ( -25.90) >DroWil_CAF1 92831 94 - 1 CCAUGUCUGGUUAACC------CCCC----GGGGCGCCAGU--------UGCUCAUGCGCAUUGUAUUUCGUGUGAAAAUCACAAUUAAAUGCGAUUUUUAUGCGCAC--CU--AC .((...(((((...((------(...----.))).))))).--------))....((((((((((((((..((((.....))))...))))))))......)))))).--..--.. ( -29.00) >DroMoj_CAF1 109667 92 - 1 CGGCUC------------GGCAACCAGCAACUGGCGCCAGU--------UGCUCAUGCGCUUUGUGUUUCGUGUGAAAAUCACAAUUAAAUGCAAUUUUUAUGCGCAC--CU--AC ......------------((.....((((((((....))))--------))))..(((((.((((((((..((((.....))))...)))))))).......))))))--).--.. ( -28.90) >DroAna_CAF1 101876 99 - 1 CGCCUC-----------UGCUAAGCA----GCGGCGCCAGUGCCAGCGCUGCUCAUGCGCAUUGUAUUUCGUGUGAAAAUCACAAUUAAAUGCGAUUUUUAUGCGCAC--CUCGCC .(((.(-----------(((...)))----).)))((..((((((((((.......))))(((((((((..((((.....))))...))))))))).....)).))))--...)). ( -35.70) >DroPer_CAF1 109885 94 - 1 CCCCCUC----------UGUCAAGCA----GCGGCGCCAGU--------UGCUCAUGCGCAUUGUAUUUCGUGUGAAAAUCACAAUUAAAUGCGAUUUUUAUGCGCACCCCUCGCC .......----------......(((----(((((....))--------))))..((((((((((((((..((((.....))))...))))))))......))))))......)). ( -25.90) >consensus CCCCUUC__________UGUCAACCA____GCGGCGCCAGU________UGCUCAUGCGCAUUGUAUUUCGUGUGAAAAUCACAAUUAAAUGCGAUUUUUAUGCGCAC__CUCGCC .................................((((.............((....))..(((((((((..((((.....))))...)))))))))......)))).......... (-18.74 = -18.63 + -0.11)

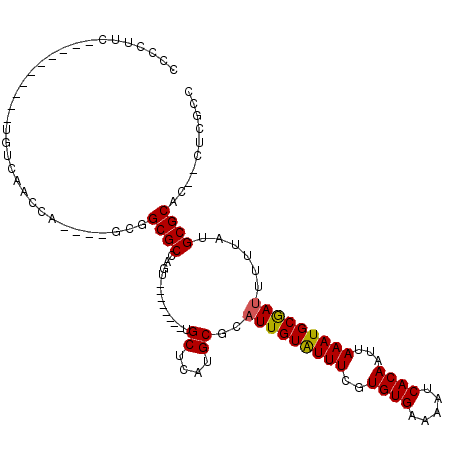
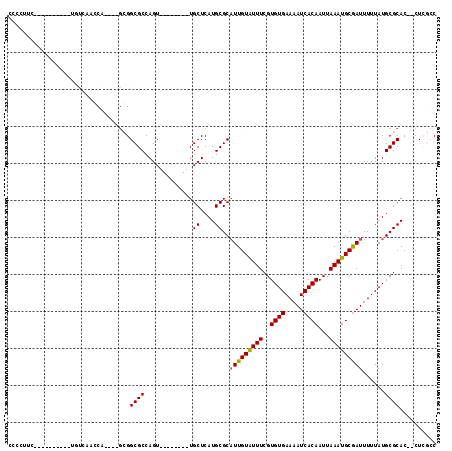
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:47 2006