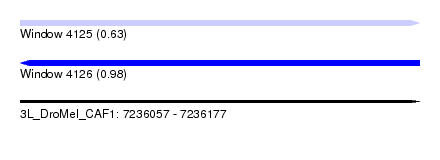
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,236,057 – 7,236,177 |
| Length | 120 |
| Max. P | 0.978592 |

| Location | 7,236,057 – 7,236,177 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -29.49 |
| Consensus MFE | -23.12 |
| Energy contribution | -23.16 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7236057 120 + 23771897 UCAUUGAUCCAUUUGGUAUUGGCAUUUGUACUGGUCUCACUAUCGCCUAAAGGCAAUUCCCCAGAUUGCAGUCUUGCUGCCAAAUGACCAAAGAGUGGCCUUUUCACUCGAAUUUCCAAC ............((((..((((((((((..((((.......((.(((....))).))...))))...((((.....)))))))))).)))).((((((.....))))))......)))). ( -31.70) >DroSec_CAF1 17611 119 + 1 UCAUUGAUCCAUUUGGUAUUGGCAUGUGUACUGGUCUCCCUGUCGCCUAAAGGCAAUUCCACAGAUUGCAGUCUUGCUGCCAAAUGACCAAAGAGUG-CCUUUUCCCUCGAAUUGCCAAC ............((((((..(((((......(((((...((((.(((....)))......))))...((((.....)))).....)))))....)))-))..(((....))).)))))). ( -29.00) >DroSim_CAF1 17624 119 + 1 UCAUUGAUCCAUUUGGUAUUGGCAUUUGUACUGGUCUCCCUGUCGCCUAAAGGCAAUUCCACAGAUUGCAGUCUUGCUGCCAAAUGACCAAAGAGUG-CCUUUUCACUCGAAUUGCCAAC ............((((((((((((((((..((((.(.....).)(((....))).......)))...((((.....)))))))))).)))).(((((-......)))))....)))))). ( -33.00) >DroEre_CAF1 18109 116 + 1 UCAUUGAUCCAUUUGGUAUUGGCAUUUGUACUGGUCCCCCUGUUGCCUAGAGGCAAUUCCACAGAUUGCAGU-UUGCUGCCAAAUGACCAAAG--UU-GCUUUUCACUCGAAUUUCCACC ...((((.......((((((((((((((..(((........((((((....))))))....)))...((((.-...)))))))))).))))..--.)-)))......))))......... ( -26.82) >DroYak_CAF1 19164 116 + 1 UCAUUGAUCCAUUUGGUAUUGGCAUUUGUACUGGUCUCCCUGUCGCCUAAAGGCAAUUUCACAGAUUGCAGU-UUGCUGCCAAAUGACCAAAA--UG-CCUUUUCACUCGAAUUUCCACC ...((((.......((((((((((((((..((((.(.....).)(((....))).......)))...((((.-...)))))))))).)))..)--))-)).......))))......... ( -26.94) >consensus UCAUUGAUCCAUUUGGUAUUGGCAUUUGUACUGGUCUCCCUGUCGCCUAAAGGCAAUUCCACAGAUUGCAGUCUUGCUGCCAAAUGACCAAAGAGUG_CCUUUUCACUCGAAUUUCCAAC ...........((((((.((((((.....((((((((....((.(((....))).)).....))))..)))).....))))))...))))))............................ (-23.12 = -23.16 + 0.04)


| Location | 7,236,057 – 7,236,177 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -29.06 |
| Energy contribution | -30.02 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
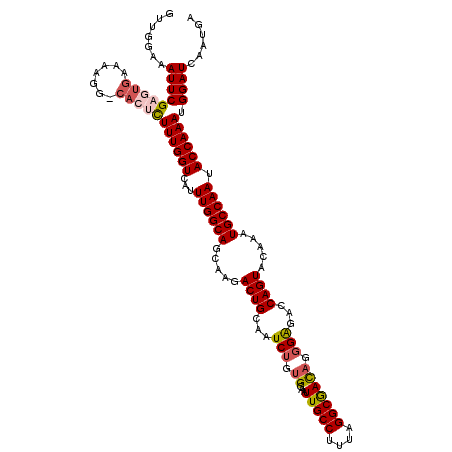
>3L_DroMel_CAF1 7236057 120 - 23771897 GUUGGAAAUUCGAGUGAAAAGGCCACUCUUUGGUCAUUUGGCAGCAAGACUGCAAUCUGGGGAAUUGCCUUUAGGCGAUAGUGAGACCAGUACAAAUGCCAAUACCAAAUGGAUCAAUGA .((((......(((((.......))))).((((.((((((((((.....))))...((((...((((((....)))))).......))))..))))))))))..))))............ ( -35.70) >DroSec_CAF1 17611 119 - 1 GUUGGCAAUUCGAGGGAAAAGG-CACUCUUUGGUCAUUUGGCAGCAAGACUGCAAUCUGUGGAAUUGCCUUUAGGCGACAGGGAGACCAGUACACAUGCCAAUACCAAAUGGAUCAAUGA ((((((..(((....)))...)-).((.((((((...(((((((((....)))....((((...(((((....)))))..((....))....)))))))))).)))))).))..)))).. ( -36.60) >DroSim_CAF1 17624 119 - 1 GUUGGCAAUUCGAGUGAAAAGG-CACUCUUUGGUCAUUUGGCAGCAAGACUGCAAUCUGUGGAAUUGCCUUUAGGCGACAGGGAGACCAGUACAAAUGCCAAUACCAAAUGGAUCAAUGA (((((((....(((((......-))))).((((((.....((((.....))))..(((.((...(((((....))))))).)))))))))......))))))).((....))........ ( -39.10) >DroEre_CAF1 18109 116 - 1 GGUGGAAAUUCGAGUGAAAAGC-AA--CUUUGGUCAUUUGGCAGCAA-ACUGCAAUCUGUGGAAUUGCCUCUAGGCAACAGGGGGACCAGUACAAAUGCCAAUACCAAAUGGAUCAAUGA ((((.......((((.......-.)--)))(((.((((((((((...-.))))...........(((((....)))))..((....))....))))))))).)))).............. ( -30.80) >DroYak_CAF1 19164 116 - 1 GGUGGAAAUUCGAGUGAAAAGG-CA--UUUUGGUCAUUUGGCAGCAA-ACUGCAAUCUGUGAAAUUGCCUUUAGGCGACAGGGAGACCAGUACAAAUGCCAAUACCAAAUGGAUCAAUGA ((((....(((....)))..((-((--((((((((.....((((...-.))))..(((.((...(((((....))))))).))))))))....)))))))..)))).............. ( -34.10) >consensus GUUGGAAAUUCGAGUGAAAAGG_CACUCUUUGGUCAUUUGGCAGCAAGACUGCAAUCUGUGGAAUUGCCUUUAGGCGACAGGGAGACCAGUACAAAUGCCAAUACCAAAUGGAUCAAUGA .......(((((((((.......)))))((((((...((((((.....((((...(((.((...(((((....))))))).)))...)))).....)))))).)))))).))))...... (-29.06 = -30.02 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:01 2006