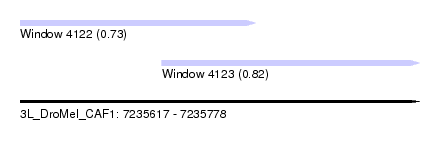
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,235,617 – 7,235,778 |
| Length | 161 |
| Max. P | 0.824600 |

| Location | 7,235,617 – 7,235,712 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.76 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -25.05 |
| Energy contribution | -25.65 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7235617 95 + 23771897 AAAACUAGAAACACUUGGCCGCUGGCGUUGGUUUUGAAUUUCUAGCUG---------------CAACUUUUA----------UUGGAUGGCCAGCGUGCUAAGCAUGGCUGGCUUUUAAG ((((((((..(..((((((((((((((((((..........)))))..---------------((.((....----------..)).))))))))).))))))..)..)))).))))... ( -30.70) >DroSec_CAF1 17168 95 + 1 AAAACUAGAAACACUUGGCCGCUGGCGUUGGUUUUGAAUUUCUAGCUG---------------CAACUUUUA----------UUGGAUGGCCAGAGUGCUAAGCAUGGCUGGCUUUUAAG ((((((((..(..(((((((.((((((((((..........)))))..---------------((.((....----------..)).))))))).).))))))..)..)))).))))... ( -25.00) >DroSim_CAF1 17170 95 + 1 AAAACUAGAAACACUUGGCCGCUGGCGUUGGUUUUGAAUUUCUAGCUG---------------CAACUUUUA----------UUGGAUGGCCAGCGUGCUAAGCAUGGCUGGCUUUUAAG ((((((((..(..((((((((((((((((((..........)))))..---------------((.((....----------..)).))))))))).))))))..)..)))).))))... ( -30.70) >DroEre_CAF1 17665 110 + 1 AAAACUAGAAACACUUGGCCGCUGGCGUUGGUUUUGAAUUUCUAGCUGCCGCUGCUGUCCGUGCAACUUUUA----------UUGGAUGGCAAGCGUGCUAAGCAUGGCUGGCUUUUAAA ((((((((..(..((((((((((((((((((..........))))).)))..(((((((((...........----------.))))))))))))).))))))..)..)))).))))... ( -36.80) >DroYak_CAF1 18732 120 + 1 AAAACUAGAAACACUUGGCCGCUGGCGUUGGUUUUGAAUUUCUAGCUGCCGCCGCUGUCCGUGCAACUUUUAUUGGAUUUUAUUGGAUGGCAAGCGUGCUAAGCAUGGCUGGCUUUUAAG ((((((((..(..((((((((((((((..((((..(.....).))))..))))((((((((.....((......)).......)))))))).)))).))))))..)..)))).))))... ( -37.60) >consensus AAAACUAGAAACACUUGGCCGCUGGCGUUGGUUUUGAAUUUCUAGCUG_______________CAACUUUUA__________UUGGAUGGCCAGCGUGCUAAGCAUGGCUGGCUUUUAAG ((((((((..(..((((((((((((((((((..........))))).................((.((................)).))))))))).))))))..)..)))).))))... (-25.05 = -25.65 + 0.60)

| Location | 7,235,674 – 7,235,778 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -26.69 |
| Consensus MFE | -20.85 |
| Energy contribution | -21.65 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7235674 104 + 23771897 --UUGGAUGGCCAGCGUGCUAAGCAUGGCUGGCUUUUAAGCAUAAACAUGGCCAACGGCGAAGACACGAAGAAAUAGUGCCGAAG--------------AAAAAUCCAACCAACUGAACA --(((((((((((.(((((...)))))(((........))).......)))))..(((((.................)))))...--------------....))))))........... ( -27.93) >DroSec_CAF1 17225 104 + 1 --UUGGAUGGCCAGAGUGCUAAGCAUGGCUGGCUUUUAAGCAUAAACAUGGCCAACGGCGAAGACACGAUGAAAUAGUGCCGAAG--------------AAAAAUCCAACCAACUGAACA --(((((((((((..(((((((((.......))))...))))).....)))))..(((((.................)))))...--------------....))))))........... ( -27.53) >DroSim_CAF1 17227 104 + 1 --UUGGAUGGCCAGCGUGCUAAGCAUGGCUGGCUUUUAAGCAUAAACAUGGCCAACGGCGAAGACACGAUGAAAUAGUGCCGAAG--------------AAAAAUCCAACCAACUGAACA --(((((((((((.(((((...)))))(((........))).......)))))..(((((.................)))))...--------------....))))))........... ( -27.93) >DroEre_CAF1 17737 102 + 1 --UUGGAUGGCAAGCGUGCUAAGCAUGGCUGGCUUUUAAACAUAAACAUCGCCAGCGGCGAAGACACGAAAAAAUAAUGCCGAAG--------------AAAAAUC--ACCAACUGAACA --((((.(((((..((((((..((...((((((.................)))))).))..)).)))).........)))))..(--------------(....))--.))))....... ( -25.73) >DroYak_CAF1 18812 118 + 1 UAUUGGAUGGCAAGCGUGCUAAGCAUGGCUGGCUUUUAAGCAUAAACAUCGCCAGCGGCGAAGACACGAAAAAAUAAUACCGAAGACAGAUGAAAAAUGAAAAACC--ACCAACUGAACA ..((((.(((....((((((..((...((((((.................)))))).))..)).))))..................((..(....).)).....))--)))))....... ( -24.33) >consensus __UUGGAUGGCCAGCGUGCUAAGCAUGGCUGGCUUUUAAGCAUAAACAUGGCCAACGGCGAAGACACGAAGAAAUAGUGCCGAAG______________AAAAAUCCAACCAACUGAACA ..((((.((((((..(((((((((.......))))...))))).....)))))).(((((.................)))))...........................))))....... (-20.85 = -21.65 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:58 2006