
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,235,322 – 7,235,544 |
| Length | 222 |
| Max. P | 0.875366 |

| Location | 7,235,322 – 7,235,424 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.14 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -14.12 |
| Energy contribution | -13.88 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7235322 102 + 23771897 AACCGCAACAGCCGCA---------GCAGGAUUACGUAACUUUCCCUGUAUCUUGCGAAUUACGCUGCUGACCAUGAGCAUCGGUUUUUAGCUGAUAGCUGAAAAG---------CUAUC ....((..((((.((.---------(((((((.(((..........))))))))))(.....))).))))(((.........))).....)).(((((((....))---------))))) ( -28.10) >DroSec_CAF1 16875 102 + 1 AACCGCAAGAGCCGCA---------GCAGCAUUACGUAACUUUCCCUGCAUCUUGCGAAUUACGCUGCUGACCAUGAGCAGUGGCUUUUUUCUUUUAGCUGAAAAG---------CUAUC ...(((((((...(((---------(...................)))).))))))).....(((((((.......)))))))............(((((....))---------))).. ( -30.21) >DroSim_CAF1 16891 102 + 1 AACCGCAACAGCCGCA---------GCAGCAUUACGUAACUUUCCCUGCAUCUUGCGAAUUACGCUGCUGACCAUGAGCAGUGGCUUUUUUUCUUUAGCUAAAAAG---------CUAUC ....((..((....((---------((((((((.(((((.............))))))))...)))))))....)).)).((((((((((..(....)..))))))---------)))). ( -25.02) >DroEre_CAF1 17390 105 + 1 AACCGCAGCAGCCACAGCCACUGCCGCAGCAUUACGUAACUUUCGCUGCAUCUUGCGAAUUACGCUGCUGACCAUGAGCAGUGGCUUUU------CAGCUGAAAAG---------UUAUC (((..((((......((((((((((((((((...(((((..(((((........)))))))))).)))))....)).)))))))))...------..))))....)---------))... ( -37.50) >DroYak_CAF1 18445 102 + 1 AACCGCAACAG------------CCGCAGCAUUACGUAACUUUCCCUGCAUCUUGCGAAUUACGCUGCUGACCAUGAGCACUGGCUUUU------CAGCUGCAAACUUACCAAACUUAUC ...(((((.(.------------..((((................)))).).)))))......((.(((((....((((....)))).)------)))).)).................. ( -21.09) >consensus AACCGCAACAGCCGCA_________GCAGCAUUACGUAACUUUCCCUGCAUCUUGCGAAUUACGCUGCUGACCAUGAGCAGUGGCUUUU______UAGCUGAAAAG_________CUAUC ...(((((.................((((................))))...))))).....(((((((.......)))))))(((..........)))..................... (-14.12 = -13.88 + -0.24)

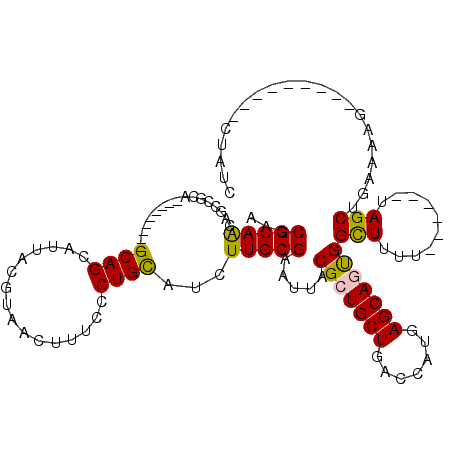

| Location | 7,235,353 – 7,235,464 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.11 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.52 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7235353 111 + 23771897 UUUCCCUGUAUCUUGCGAAUUACGCUGCUGACCAUGAGCAUCGGUUUUUAGCUGAUAGCUGAAAAG---------CUAUCUUUGCCAAAACUCUAAUUGUUACCGAAUUCGGCGCAAUCA ............((((((((((((.((((.......)))).))(((((..((.(((((((....))---------)))))...)).)))))..)))))....(((....))))))))... ( -29.20) >DroSec_CAF1 16906 111 + 1 UUUCCCUGCAUCUUGCGAAUUACGCUGCUGACCAUGAGCAGUGGCUUUUUUCUUUUAGCUGAAAAG---------CUAUCAUCGCCAAAACUCUAAUUGUUACCGAAUUCGGCGCAAUCA ......(((.....((((....(((((((.......)))))))............(((((....))---------)))...)))).................(((....))).))).... ( -28.10) >DroSim_CAF1 16922 111 + 1 UUUCCCUGCAUCUUGCGAAUUACGCUGCUGACCAUGAGCAGUGGCUUUUUUUCUUUAGCUAAAAAG---------CUAUCAUCGCCAAAACUCUAAUUGUUACCGAAUUCGGCGCAAUCA ......(((.....((((....(((((((.......)))))))(((((((..(....)..))))))---------).....)))).................(((....))).))).... ( -26.90) >DroEre_CAF1 17430 105 + 1 UUUCGCUGCAUCUUGCGAAUUACGCUGCUGACCAUGAGCAGUGGCUUUU------CAGCUGAAAAG---------UUAUCAACGCCAAAACUCUAAUUGUUGCCGAAUUCGGCGCAAUCA .(((((........)))))...(((((((.......)))))))((((((------(....))))))---------)...................(((((.((((....))))))))).. ( -31.60) >DroYak_CAF1 18473 114 + 1 UUUCCCUGCAUCUUGCGAAUUACGCUGCUGACCAUGAGCACUGGCUUUU------CAGCUGCAAACUUACCAAACUUAUCAUCGCCAAAACUCUAAUUGUUACCGAAUUCGGCGCAAUCA ............(((((......((.(((((....((((....)))).)------)))).))........................................(((....))))))))... ( -22.40) >consensus UUUCCCUGCAUCUUGCGAAUUACGCUGCUGACCAUGAGCAGUGGCUUUU______UAGCUGAAAAG_________CUAUCAUCGCCAAAACUCUAAUUGUUACCGAAUUCGGCGCAAUCA ......(((.....((((....(((((((.......)))))))(((..........)))......................)))).................(((....))).))).... (-18.56 = -18.52 + -0.04)

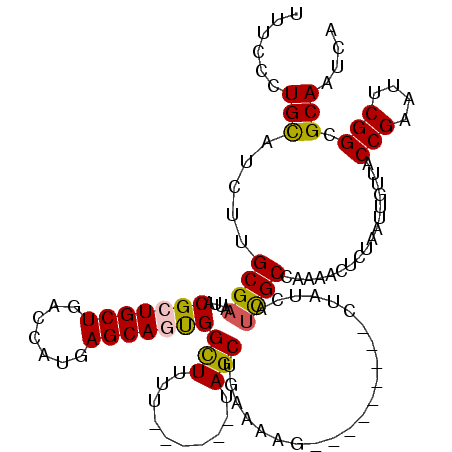

| Location | 7,235,424 – 7,235,544 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.03 |
| Mean single sequence MFE | -15.88 |
| Consensus MFE | -11.06 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7235424 120 + 23771897 UUUGCCAAAACUCUAAUUGUUACCGAAUUCGGCGCAAUCACGAAACUCAUAACUUCAUGUUGACCCCAAACCAAAAAUAAAAAAUAAAAACGAAAAAAAAACCCCAGCACAAUAAGAGAC ..........((((.(((((..(((....))).((......(.....).((((.....))))............................................))))))).)))).. ( -13.50) >DroSec_CAF1 16977 111 + 1 AUCGCCAAAACUCUAAUUGUUACCGAAUUCGGCGCAAUCACGAAACUCACAACUUCAUGUUGACCCCAAACCAAAAAUAAAAACC---------AGAAAAACCCCAGCACAAUAAGAGAC ..........((((.(((((..(((....))).((......(.....).((((.....)))).......................---------............))))))).)))).. ( -15.60) >DroSim_CAF1 16993 98 + 1 AUCGCCAAAACUCUAAUUGUUACCGAAUUCGGCGCAAUCACGAAACUCACAACUUCAUGUUGACCCCAAA-------------CG---------AGAAAAACCCCAGCACAAUAAGAGAC ..........((((.(((((..(((....))).((......(.....).((((.....))))........-------------..---------............))))))).)))).. ( -15.60) >DroEre_CAF1 17495 112 + 1 AACGCCAAAACUCUAAUUGUUGCCGAAUUCGGCGCAAUCACGACAAACACAACUCCAUGCUGACCCCAAAGCAAACGAAAA--------AUGUAAAAAAUACCCCGGCACAAUAAAAGAC ...(((.........(((((.((((....)))))))))...................((((........))))........--------................)))............ ( -19.80) >DroYak_CAF1 18547 109 + 1 AUCGCCAAAACUCUAAUUGUUACCGAAUUCGGCGCAAUCACGACCAAAACAACUCUAUGUUGACCCCAACUGAAAAAAAAA-----------GGAAAAAUACCCCGGCACAAUAAAAGAC ...(((.........(((((..(((....))).))))).....((........((...((((....)))).))........-----------))...........)))............ ( -14.89) >consensus AUCGCCAAAACUCUAAUUGUUACCGAAUUCGGCGCAAUCACGAAACUCACAACUUCAUGUUGACCCCAAACCAAAAAAAAA_____________AAAAAAACCCCAGCACAAUAAGAGAC ..........((((.(((((..(((....))).................((((.....))))..............................................))))).)))).. (-11.06 = -11.50 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:57 2006