
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,234,649 – 7,235,000 |
| Length | 351 |
| Max. P | 0.996850 |

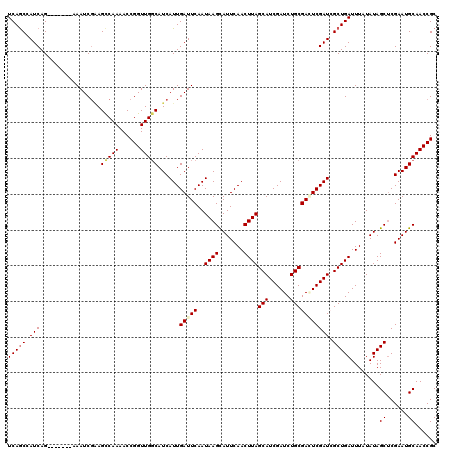
| Location | 7,234,649 – 7,234,769 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.09 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -22.34 |
| Energy contribution | -22.94 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7234649 120 + 23771897 UCAGCCAUCAGAAAUCAGAAAUCGGAGCCAAAACCGGUUGGCAUCAUUGAUUCAAUAAGCAUUCAACUUAGCAUCGAUCUGCGAGUCGAUAGCUGAUUUAUAUAGCACGAAUGCAACCGC ...((.((...(((((((..((((..(((((......)))))..)...(((((..((((.......))))(((......)))))))))))..)))))))..)).)).............. ( -27.80) >DroSec_CAF1 16218 113 + 1 UCAGCCAUCAG-------AAAUCGAAGUCAAAACCGGUUGGCAUCAUUGAUUCAAUAAGUAUUCAACUUAGCAUCGAUCUGCGACUCGAUAGCUGAUUUAUAUAGCUCGAAUGCAACCGC ...((.(((((-------..(((((.(((..((....)).(((..((((((....(((((.....)))))..)))))).)))))))))))..))))).......((......))....)) ( -24.00) >DroSim_CAF1 16215 113 + 1 UCAGCCAUCAG-------AAAUCGGAGCCAAAACCGGUUGGCAUCAUUGAUUCAAUAAGCAUUCAACUUAGCAUCGAUCUGCGACUCGAUCGCUGAUUUAUAUAGCUCGAAUGCAACCGC ...........-------....(((.(((((......)))))................((((((...(((..((.((((.((((.....)))).)))).)).)))...))))))..))). ( -25.10) >DroEre_CAF1 16735 113 + 1 UCAGCAAUCAG-------AAAUCGCAGCCAAAACCGGUUGCCAUCAUUGAUUCAAUAAGCAUUCAACUUAGCAUCGAUCUGCGAAUCGAUCGCUGAUUUAUAUAGCUCGAAUGCAACCGC ...((((((((-------..((.((((((......)))))).))..))))))......((((((((.(((((((((((......)))))).))))).)).........))))))....)) ( -29.70) >DroYak_CAF1 17770 113 + 1 UCAACAAUCAG-------AAAUCCAAGCCAAAACCGGUUGGCAUCAUUGAUUCAAUAAGCAUUCAACUUAGCAUCGAUCUGCGAGUCGAUCGCUGAUUUAUAUAGCUCGAAUGCAACCGC .....((((((-------..(((...(((((......)))))......(((((..((((.......))))(((......)))))))))))..))))))......((......))...... ( -24.90) >consensus UCAGCCAUCAG_______AAAUCGAAGCCAAAACCGGUUGGCAUCAUUGAUUCAAUAAGCAUUCAACUUAGCAUCGAUCUGCGACUCGAUCGCUGAUUUAUAUAGCUCGAAUGCAACCGC (((((.(((.................(((((......)))))......(((((..((((.......))))(((......))))))))))).)))))........((......))...... (-22.34 = -22.94 + 0.60)


| Location | 7,234,769 – 7,234,881 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.19 |
| Mean single sequence MFE | -21.28 |
| Consensus MFE | -17.02 |
| Energy contribution | -16.74 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7234769 112 - 23771897 AUGUCAUCAUCACAUAUAUGUAAUUAAUGUCUCGUGUUGCUUGCACAUUCAUUGCGUGCAAAUUAUACGAAAAAGUAUCCGACAUUCAAACAUUCAGAC--------AUUCAGGCAUUCC (((((......(((....)))....((((((((((((...((((((.........))))))...))))))....((.....)).............)))--------)))..)))))... ( -20.90) >DroSec_CAF1 16331 108 - 1 AUGUCAUCAUCACA----UGUAAUUAAUGCCUCGUGUCGCUUGCACAUUCAUUGCGUGCAAAUUAUACGAAAAAGUAUCCGACAUUCAAACAUUCAGAC--------AUUCAGGCAUUCC ..............----.......(((((((.(((((..((((((.........))))))...((((......))))..................)))--------))..))))))).. ( -22.10) >DroSim_CAF1 16328 108 - 1 AUGUCAUCAUCACA----UGUAAUUAAUGCCUCGUGUCGCUUGCACAUUCAUUGCGUGCAAAUUAUACGAAAAAGUAUCCGACAUUCAAACAUUCAGAC--------AUUCAGGCAUUCC ..............----.......(((((((.(((((..((((((.........))))))...((((......))))..................)))--------))..))))))).. ( -22.10) >DroEre_CAF1 16848 116 - 1 AUGUCAUCAUCACA----CGUAAUUAAUGCCUCGUGUCGCUUGCACAUUCAUUGCGUGCAUAUUAUACAAAAAAGUAUCCGACAUUCAGACAUUCAAACAUUCAUACACUCAGGCAUUCC ..............----.......(((((((.((((((..(((((.........)))))....((((......)))).))))))...(.....)................))))))).. ( -18.60) >DroYak_CAF1 17883 116 - 1 AUGUCAUCAUCACA----CGUAAUUAAUGCCUCGUGUCGCUUGCACAUUCAUUGCGUGCAAAUUAUACAAAAAAGUAUCCGACACUCAGACAUUCAGACAGUCAAAGAUUCAGACAUUCC (((((...(((...----...............((((((.((((((.........))))))...((((......)))).))))))...(((.........)))...)))...)))))... ( -22.70) >consensus AUGUCAUCAUCACA____UGUAAUUAAUGCCUCGUGUCGCUUGCACAUUCAUUGCGUGCAAAUUAUACGAAAAAGUAUCCGACAUUCAAACAUUCAGAC________AUUCAGGCAUUCC (((((......((......))....((((....((((((.((((((.........))))))...((((......)))).)))))).....))))..................)))))... (-17.02 = -16.74 + -0.28)

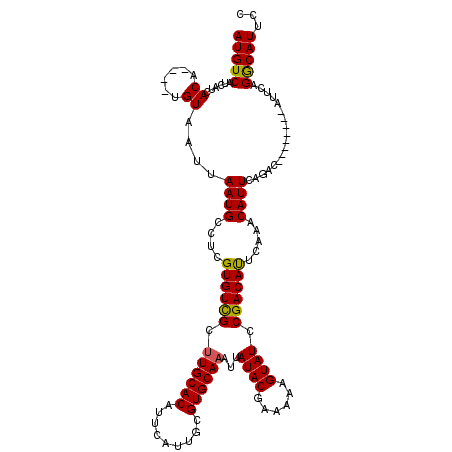

| Location | 7,234,801 – 7,234,920 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.86 |
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -26.50 |
| Energy contribution | -25.94 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7234801 119 + 23771897 GGAUACUUUUUCGUAUAAUUUGCACGCAAUGAAUGUGCAAGCAACACGAGACAUUAAUUACAUAUAUGUGAUGAUGACAUCACCAUCACCAAGUCAUCUGUUGGUUUGCAGAUU-UCAAU .(((....((((((....((((((((.......))))))))....))))))................((((((.((....)).))))))...)))((((((......)))))).-..... ( -27.40) >DroSec_CAF1 16363 115 + 1 GGAUACUUUUUCGUAUAAUUUGCACGCAAUGAAUGUGCAAGCGACACGAGGCAUUAAUUACA----UGUGAUGAUGACAUCACCAUCACCAAGUCAUCUGUCGGCUUGCAGAUU-UCAAU ..((((......))))...(((....)))(((((.(((((((((((.(((((..........----.((((((.((....)).))))))...))).)))))).))))))).).)-))).. ( -28.84) >DroSim_CAF1 16360 115 + 1 GGAUACUUUUUCGUAUAAUUUGCACGCAAUGAAUGUGCAAGCGACACGAGGCAUUAAUUACA----UGUGAUGAUGACAUCACCAUCACCAAGUCAUCUGCCGGUUUGCAGAUU-UCAAU .(((....((((((.(..((((((((.......))))))))..).))))))...........----.((((((.((....)).))))))...)))((((((......)))))).-..... ( -28.00) >DroEre_CAF1 16888 116 + 1 GGAUACUUUUUUGUAUAAUAUGCACGCAAUGAAUGUGCAAGCGACACGAGGCAUUAAUUACG----UGUGAUGAUGACAUCACCAGCACCAAGUCAUCUGUCGGCUUGCAGAUUUUCAAU ..((((......))))..............((((.(((((((((((.(((((.........(----.((((((....)))))))........))).)))))).))))))).))))..... ( -28.93) >DroYak_CAF1 17923 116 + 1 GGAUACUUUUUUGUAUAAUUUGCACGCAAUGAAUGUGCAAGCGACACGAGGCAUUAAUUACG----UGUGAUGAUGACAUCAUCAUCAGCAAGUCAUCUGUCGGUUUGCAGAUUUUCAAU ..........(((...((((((((((((((((...(((.....(((((............))----)))(((((((....))))))).)))..)))).)).))...))))))))..))). ( -30.90) >consensus GGAUACUUUUUCGUAUAAUUUGCACGCAAUGAAUGUGCAAGCGACACGAGGCAUUAAUUACA____UGUGAUGAUGACAUCACCAUCACCAAGUCAUCUGUCGGUUUGCAGAUU_UCAAU ..((((......)))).............(((((.(((((((((((.(((((....((((((....))))))((((.......)))).....))).)))))).))))))).))..))).. (-26.50 = -25.94 + -0.56)


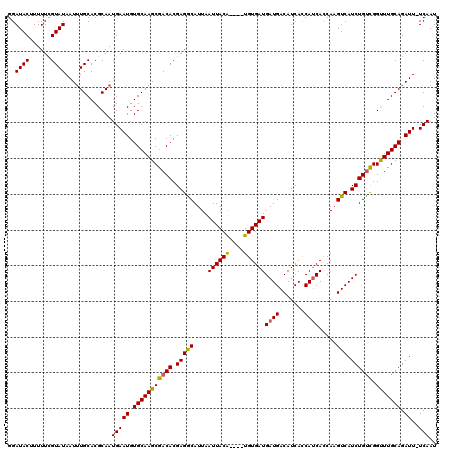
| Location | 7,234,801 – 7,234,920 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.86 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -26.66 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7234801 119 - 23771897 AUUGA-AAUCUGCAAACCAACAGAUGACUUGGUGAUGGUGAUGUCAUCAUCACAUAUAUGUAAUUAAUGUCUCGUGUUGCUUGCACAUUCAUUGCGUGCAAAUUAUACGAAAAAGUAUCC .....-.(((((........))))).((((.(((((((((....)))))))))..................((((((...((((((.........))))))...))))))..)))).... ( -31.40) >DroSec_CAF1 16363 115 - 1 AUUGA-AAUCUGCAAGCCGACAGAUGACUUGGUGAUGGUGAUGUCAUCAUCACA----UGUAAUUAAUGCCUCGUGUCGCUUGCACAUUCAUUGCGUGCAAAUUAUACGAAAAAGUAUCC ..(((-(...(((((((.((((((.......(((((((((....))))))))).----.(((.....))).)).)))))))))))..))))(((....)))...((((......)))).. ( -31.30) >DroSim_CAF1 16360 115 - 1 AUUGA-AAUCUGCAAACCGGCAGAUGACUUGGUGAUGGUGAUGUCAUCAUCACA----UGUAAUUAAUGCCUCGUGUCGCUUGCACAUUCAUUGCGUGCAAAUUAUACGAAAAAGUAUCC .....-.((((((......)))))).((((.(((((((((....))))))))).----.............((((((.(.((((((.........)))))).).))))))..)))).... ( -34.60) >DroEre_CAF1 16888 116 - 1 AUUGAAAAUCUGCAAGCCGACAGAUGACUUGGUGCUGGUGAUGUCAUCAUCACA----CGUAAUUAAUGCCUCGUGUCGCUUGCACAUUCAUUGCGUGCAUAUUAUACAAAAAAGUAUCC ..((((....(((((((.((((((.(..(((((....((((((....)))))).----....)))))..).)).)))))))))))..)))).............((((......)))).. ( -27.50) >DroYak_CAF1 17923 116 - 1 AUUGAAAAUCUGCAAACCGACAGAUGACUUGCUGAUGAUGAUGUCAUCAUCACA----CGUAAUUAAUGCCUCGUGUCGCUUGCACAUUCAUUGCGUGCAAAUUAUACAAAAAAGUAUCC .......(((((........))))).((((((.(((((((....)))))))(((----((............))))).))((((((.........))))))...........)))).... ( -26.10) >consensus AUUGA_AAUCUGCAAACCGACAGAUGACUUGGUGAUGGUGAUGUCAUCAUCACA____UGUAAUUAAUGCCUCGUGUCGCUUGCACAUUCAUUGCGUGCAAAUUAUACGAAAAAGUAUCC .......(((((........))))).((((.(((((((((....)))))))))..................((((((...((((((.........))))))...))))))..)))).... (-26.66 = -27.50 + 0.84)

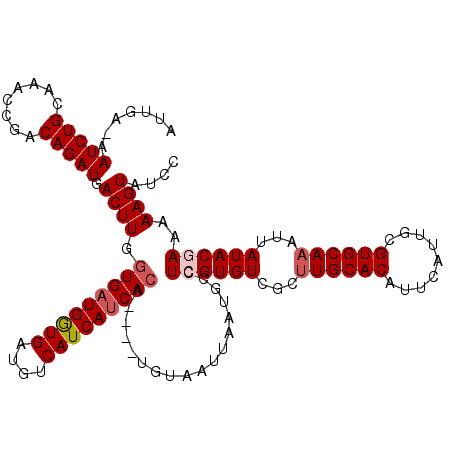

| Location | 7,234,841 – 7,234,960 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.48 |
| Mean single sequence MFE | -31.61 |
| Consensus MFE | -26.30 |
| Energy contribution | -26.06 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7234841 119 + 23771897 GCAACACGAGACAUUAAUUACAUAUAUGUGAUGAUGACAUCACCAUCACCAAGUCAUCUGUUGGUUUGCAGAUU-UCAAUUGCAUGCAGUCGCCUCAAAUGUCAGAUGAAAGUUGCAGUU (((((......................((((((.((....)).))))))....(((((((...(((((..((((-.((......)).))))....)))))..)))))))..))))).... ( -29.90) >DroSec_CAF1 16403 115 + 1 GCGACACGAGGCAUUAAUUACA----UGUGAUGAUGACAUCACCAUCACCAAGUCAUCUGUCGGCUUGCAGAUU-UCAAUUGCAUACAGUCGCCUCAAAUGUCAGAUGAAAGUUGCAGUU (((((..(((((..........----.((((((.((....)).)))))).............(((((((((...-....)))))...))))))))).....((....))..))))).... ( -31.20) >DroSim_CAF1 16400 115 + 1 GCGACACGAGGCAUUAAUUACA----UGUGAUGAUGACAUCACCAUCACCAAGUCAUCUGCCGGUUUGCAGAUU-UCAAUUGCAUACAGUCGCCUCAAAUGUCAGAUGAAAGUUGCAGUU (((((..(((((..........----.((((((.((....)).))))))...(..((((((......)))))).-.)..............))))).....((....))..))))).... ( -32.50) >DroEre_CAF1 16928 113 + 1 GCGACACGAGGCAUUAAUUACG----UGUGAUGAUGACAUCACCAGCACCAAGUCAUCUGUCGGCUUGCAGAUUUUCAAUUGCAUACAGU---CUCGAAUGUCAGAUGAAAGUUGCAGUU (((((.((((((.........(----.((((((....))))))).(((.......((((((......)))))).......))).....))---))))....((....))..))))).... ( -31.04) >DroYak_CAF1 17963 113 + 1 GCGACACGAGGCAUUAAUUACG----UGUGAUGAUGACAUCAUCAUCAGCAAGUCAUCUGUCGGUUUGCAGAUUUUCAAUUGCAUACAGU---CUCAAAUGUCAGAUGAAAGUUGCAGUU (((((..(((((..........----..((((((((....))))))))((((...((((((......))))))......)))).....))---))).....((....))..))))).... ( -33.40) >consensus GCGACACGAGGCAUUAAUUACA____UGUGAUGAUGACAUCACCAUCACCAAGUCAUCUGUCGGUUUGCAGAUU_UCAAUUGCAUACAGUCGCCUCAAAUGUCAGAUGAAAGUUGCAGUU (((((..((..(((((.(((((....))))))))))...))............(((((((.((((.(((((........))))).)).(......)...)).)))))))..))))).... (-26.30 = -26.06 + -0.24)



| Location | 7,234,841 – 7,234,960 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.48 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -26.54 |
| Energy contribution | -27.42 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7234841 119 - 23771897 AACUGCAACUUUCAUCUGACAUUUGAGGCGACUGCAUGCAAUUGA-AAUCUGCAAACCAACAGAUGACUUGGUGAUGGUGAUGUCAUCAUCACAUAUAUGUAAUUAAUGUCUCGUGUUGC ....(((((..(((((((.....((((....)).))((((.....-....))))......)))))))....(((((((((....)))))))))......................))))) ( -31.40) >DroSec_CAF1 16403 115 - 1 AACUGCAACUUUCAUCUGACAUUUGAGGCGACUGUAUGCAAUUGA-AAUCUGCAAGCCGACAGAUGACUUGGUGAUGGUGAUGUCAUCAUCACA----UGUAAUUAAUGCCUCGUGUCGC .................(((((..((((((.((((.((((.....-....)))).....))))...((...(((((((((....))))))))).----.))......))))))))))).. ( -34.20) >DroSim_CAF1 16400 115 - 1 AACUGCAACUUUCAUCUGACAUUUGAGGCGACUGUAUGCAAUUGA-AAUCUGCAAACCGGCAGAUGACUUGGUGAUGGUGAUGUCAUCAUCACA----UGUAAUUAAUGCCUCGUGUCGC .................(((((..((((((...((.((((.....-.((((((......))))))......(((((((((....))))))))).----))))))...))))))))))).. ( -37.20) >DroEre_CAF1 16928 113 - 1 AACUGCAACUUUCAUCUGACAUUCGAG---ACUGUAUGCAAUUGAAAAUCUGCAAGCCGACAGAUGACUUGGUGCUGGUGAUGUCAUCAUCACA----CGUAAUUAAUGCCUCGUGUCGC ...((((..(((((..((.(((.((..---..)).)))))..)))))...))))...(((((((.(..(((((....((((((....)))))).----....)))))..).)).))))). ( -25.40) >DroYak_CAF1 17963 113 - 1 AACUGCAACUUUCAUCUGACAUUUGAG---ACUGUAUGCAAUUGAAAAUCUGCAAACCGACAGAUGACUUGCUGAUGAUGAUGUCAUCAUCACA----CGUAAUUAAUGCCUCGUGUCGC ....((((...((((((((((......---..))).((((..(....)..))))......))))))).)))).(((((((....)))))))(((----((............)))))... ( -29.30) >consensus AACUGCAACUUUCAUCUGACAUUUGAGGCGACUGUAUGCAAUUGA_AAUCUGCAAACCGACAGAUGACUUGGUGAUGGUGAUGUCAUCAUCACA____UGUAAUUAAUGCCUCGUGUCGC .................(((((..(((((..((((.((((..(....)..)))).....))))........(((((((((....)))))))))...............)))))))))).. (-26.54 = -27.42 + 0.88)


| Location | 7,234,881 – 7,235,000 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -22.45 |
| Energy contribution | -23.05 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7234881 119 - 23771897 AAACCAAACCAACUAUUAUGCAGGUACGAAUUAUCCAACCAACUGCAACUUUCAUCUGACAUUUGAGGCGACUGCAUGCAAUUGA-AAUCUGCAAACCAACAGAUGACUUGGUGAUGGUG ..((((.(((((......(((((...................)))))....(((((((.....((((....)).))((((.....-....))))......))))))).)))))..)))). ( -28.21) >DroSec_CAF1 16439 119 - 1 AAACCAAACCAACCAUUAUGCAGGUACGAAUUAUCCAACCAACUGCAACUUUCAUCUGACAUUUGAGGCGACUGUAUGCAAUUGA-AAUCUGCAAGCCGACAGAUGACUUGGUGAUGGUG ..((((.(((((......(((((...................)))))....(((((((.......((....))((.((((.....-....)))).))...))))))).)))))..)))). ( -28.81) >DroSim_CAF1 16436 119 - 1 AAACCAAACCAACCAUUAUGCAGGUACGAAUUAUCCAACCAACUGCAACUUUCAUCUGACAUUUGAGGCGACUGUAUGCAAUUGA-AAUCUGCAAACCGGCAGAUGACUUGGUGAUGGUG ...........((((((((.(((((..............(((.((((((..((..((........))..))..)).)))).))).-.((((((......)))))).))))))))))))). ( -30.10) >DroEre_CAF1 16964 116 - 1 AAACCCAG-CAACUAUUAUGCAGGUACGAACCAACCAGCCAACUGCAACUUUCAUCUGACAUUCGAG---ACUGUAUGCAAUUGAAAAUCUGCAAGCCGACAGAUGACUUGGUGCUGGUG ....((((-((.(((...(((((...................)))))....(((((((....(((.(---.((...((((..(....)..)))))))))))))))))..))))))))).. ( -30.11) >DroYak_CAF1 17999 116 - 1 AAACCAAG-CAACUAUUAUGCAGGUACGAAAUAUCCAACCAACUGCAACUUUCAUCUGACAUUUGAG---ACUGUAUGCAAUUGAAAAUCUGCAAACCGACAGAUGACUUGCUGAUGAUG ......((-(((......(((((...................)))))....((((((((((......---..))).((((..(....)..))))......))))))).)))))....... ( -23.01) >consensus AAACCAAACCAACUAUUAUGCAGGUACGAAUUAUCCAACCAACUGCAACUUUCAUCUGACAUUUGAGGCGACUGUAUGCAAUUGA_AAUCUGCAAACCGACAGAUGACUUGGUGAUGGUG ..((((.(((((......(((((...................)))))....(((((((.......((....))((.((((..(....)..)))).))...))))))).)))))..)))). (-22.45 = -23.05 + 0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:54 2006