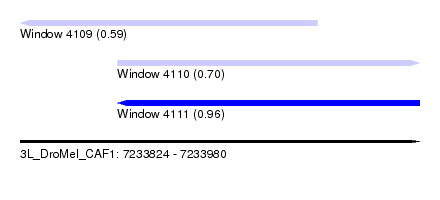
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,233,824 – 7,233,980 |
| Length | 156 |
| Max. P | 0.964991 |

| Location | 7,233,824 – 7,233,940 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.56 |
| Mean single sequence MFE | -20.96 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.58 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7233824 116 - 23771897 AAUUAUAAAAGGCAACACCCAU--CUCAUAAAUCGCUGUCUGCGUUUCGCCGCCAAAAGACCUUGACGGCCAGCCAAAAUACAUUAACUUUAUAUACAAAUAUACAAUGG-GGAGAAAA- ..........(....).(((((--.........(((.....)))....((((.(((......))).))))....................................))))-).......- ( -18.90) >DroSec_CAF1 15396 113 - 1 AAUUAUAAAAGGCAACACCCAU--CUCAUAAAUCGCUGUCUGCGUUUCGCCGCCAAAAGACCUUGACGGCCAGCCAAAAUACAAUAACUUUAUAAACAAAUAUACAAUGG-GGGGA---- ..........(....).(((..--(((((....(((.....)))....((((.(((......))).))))....................................))))-)))).---- ( -20.10) >DroSim_CAF1 15357 113 - 1 AAUUAUAAAAGGCAACACCCAU--CUCAUAAAUCGCUGUCUGCGUUUCGCCGCCAAAAGACCUUGACGGCCAGCCAAAAUACAAUAACUCUAUAAACAAAUAUACAAUGG-GGGAG---- ..........(((.........--.........(((.....)))....((((.(((......))).))))..)))............((((((.............))))-))...---- ( -20.02) >DroEre_CAF1 15809 117 - 1 AAUUAUAAAAGGCAACACCCCU--CUCAUAAAUCGCUGUCUGCGUUUUGCCGCCAAAAGACCUUGGCGGCCAGCCAAAAUACAUUAACUUUAUAUACUCGUAUACAAAUG-ACCGACAAC ..........(((.........--.....(((.(((.....))).)))((((((((......))))))))..)))......((((.....(((((....)))))..))))-......... ( -24.40) >DroYak_CAF1 16848 111 - 1 AAUUAUAAAAGGCAACACCCCUCUCUCAUAAAUCGCUGUCUGCGUUUUGCCGCCAAAAGACCUUGACGGCCAGCCAAAAUACAA-AACUUUAUACA--------CAAAUGUAUAGACAAC ..........(((................(((.(((.....))).)))((((.(((......))).))))..))).........-...((((((((--------....)))))))).... ( -21.40) >consensus AAUUAUAAAAGGCAACACCCAU__CUCAUAAAUCGCUGUCUGCGUUUCGCCGCCAAAAGACCUUGACGGCCAGCCAAAAUACAAUAACUUUAUAAACAAAUAUACAAUGG_GGAGA_AA_ ..........(((....................(((.....)))....((((.(((......))).))))..)))............................................. (-16.58 = -16.58 + 0.00)

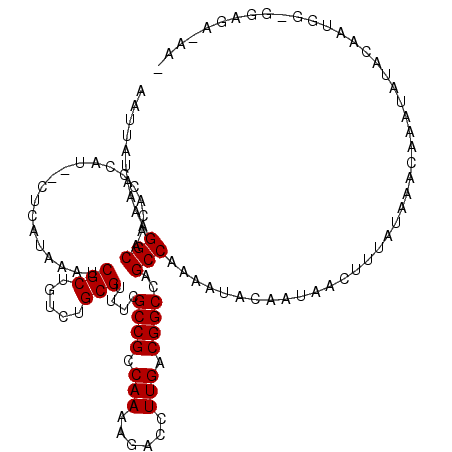
| Location | 7,233,862 – 7,233,980 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.76 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -28.68 |
| Energy contribution | -28.52 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7233862 118 + 23771897 AUUUUGGCUGGCCGUCAAGGUCUUUUGGCGGCGAAACGCAGACAGCGAUUUAUGAG--AUGGGUGUUGCCUUUUAUAAUUUGGCCAACCAACUUGAUGUUGUAUAUUAUUUGCUUUUGCU ...((((((.(((((((((....)))))))))((((.((((.((.(.(((.....)--)).).)))))).)))).......)))))).((((.....))))..........((....)). ( -32.90) >DroSec_CAF1 15431 118 + 1 AUUUUGGCUGGCCGUCAAGGUCUUUUGGCGGCGAAACGCAGACAGCGAUUUAUGAG--AUGGGUGUUGCCUUUUAUAAUUUGGCCAACCAACUUGAUGUUGUAUAUUAUUUGCUCUUGCU ...((((((.(((((((((....)))))))))((((.((((.((.(.(((.....)--)).).)))))).)))).......)))))).((((.....))))..........((....)). ( -32.90) >DroSim_CAF1 15392 118 + 1 AUUUUGGCUGGCCGUCAAGGUCUUUUGGCGGCGAAACGCAGACAGCGAUUUAUGAG--AUGGGUGUUGCCUUUUAUAAUUUGGCCAACUAACUUGAUGUUGUAUAUUAUUUGCUUUUGCU .....(..(.(((((((((....))))))))).)..)(((((.(((((..((..(.--((((((((((((...........)).))))..)))).)).)..))......)))))))))). ( -31.20) >DroEre_CAF1 15848 115 + 1 AUUUUGGCUGGCCGCCAAGGUCUUUUGGCGGCAAAACGCAGACAGCGAUUUAUGAG--AGGGGUGUUGCCUUUUAUAAUUUGGCCAAA---GCUGAUGUUGUAUAAUAUUUGCCUUUGCU ..(((((((.(((((((((....)))))))))....(((.....)))..(((((((--(((.......))))))))))...)))))))---((.((((((....)))))).))....... ( -39.70) >DroYak_CAF1 16879 117 + 1 AUUUUGGCUGGCCGUCAAGGUCUUUUGGCGGCAAAACGCAGACAGCGAUUUAUGAGAGAGGGGUGUUGCCUUUUAUAAUUUGGCCAAC---GUUGAUGUUGUAUAAUAUUUGCCUUUGCU ...((((((.(((((((((....)))))))))....(((.....)))..(((((((((.(........))))))))))...)))))).---((.((((((....)))))).))....... ( -30.90) >consensus AUUUUGGCUGGCCGUCAAGGUCUUUUGGCGGCGAAACGCAGACAGCGAUUUAUGAG__AUGGGUGUUGCCUUUUAUAAUUUGGCCAAC_AACUUGAUGUUGUAUAUUAUUUGCUUUUGCU ...((((((.(((((((((....)))))))))....(((.....)))..(((((((....(((.....))))))))))...))))))........................((....)). (-28.68 = -28.52 + -0.16)

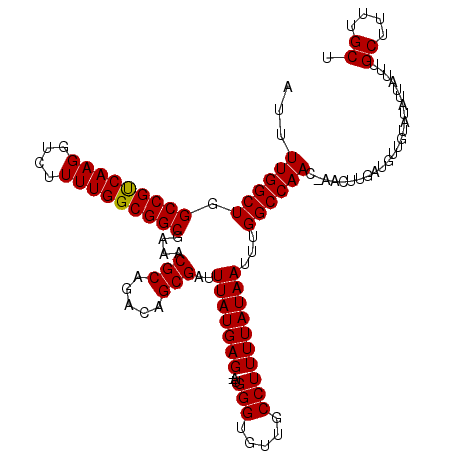
| Location | 7,233,862 – 7,233,980 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.76 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -21.12 |
| Energy contribution | -21.16 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
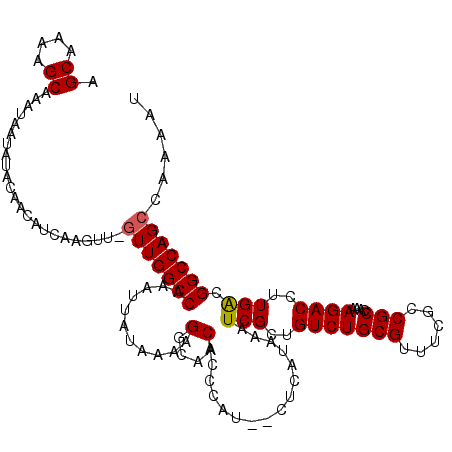
>3L_DroMel_CAF1 7233862 118 - 23771897 AGCAAAAGCAAAUAAUAUACAACAUCAAGUUGGUUGGCCAAAUUAUAAAAGGCAACACCCAU--CUCAUAAAUCGCUGUCUGCGUUUCGCCGCCAAAAGACCUUGACGGCCAGCCAAAAU .((....))....................((((((((((...........(....)......--........(((..(((((((......)))....))))..))).))))))))))... ( -28.10) >DroSec_CAF1 15431 118 - 1 AGCAAGAGCAAAUAAUAUACAACAUCAAGUUGGUUGGCCAAAUUAUAAAAGGCAACACCCAU--CUCAUAAAUCGCUGUCUGCGUUUCGCCGCCAAAAGACCUUGACGGCCAGCCAAAAU .((....))....................((((((((((...........(....)......--........(((..(((((((......)))....))))..))).))))))))))... ( -28.10) >DroSim_CAF1 15392 118 - 1 AGCAAAAGCAAAUAAUAUACAACAUCAAGUUAGUUGGCCAAAUUAUAAAAGGCAACACCCAU--CUCAUAAAUCGCUGUCUGCGUUUCGCCGCCAAAAGACCUUGACGGCCAGCCAAAAU .((....)).......................(((((((...........(....)......--........(((..(((((((......)))....))))..))).)))))))...... ( -22.30) >DroEre_CAF1 15848 115 - 1 AGCAAAGGCAAAUAUUAUACAACAUCAGC---UUUGGCCAAAUUAUAAAAGGCAACACCCCU--CUCAUAAAUCGCUGUCUGCGUUUUGCCGCCAAAAGACCUUGGCGGCCAGCCAAAAU .((.(((((..................))---))).))............(((.........--.....(((.(((.....))).)))((((((((......))))))))..)))..... ( -28.67) >DroYak_CAF1 16879 117 - 1 AGCAAAGGCAAAUAUUAUACAACAUCAAC---GUUGGCCAAAUUAUAAAAGGCAACACCCCUCUCUCAUAAAUCGCUGUCUGCGUUUUGCCGCCAAAAGACCUUGACGGCCAGCCAAAAU ......(((.....((((.((((......---))))(((...........)))..............))))...((((((...(((((........)))))...))))))..)))..... ( -24.20) >consensus AGCAAAAGCAAAUAAUAUACAACAUCAAGUU_GUUGGCCAAAUUAUAAAAGGCAACACCCAU__CUCAUAAAUCGCUGUCUGCGUUUCGCCGCCAAAAGACCUUGACGGCCAGCCAAAAU .((....)).......................(((((((...........(....)................(((..(((((((......)))....))))..))).)))))))...... (-21.12 = -21.16 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:48 2006