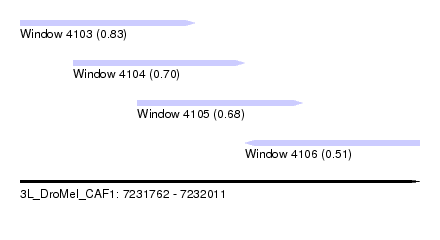
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,231,762 – 7,232,011 |
| Length | 249 |
| Max. P | 0.828956 |

| Location | 7,231,762 – 7,231,871 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.15 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -15.18 |
| Energy contribution | -17.82 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7231762 109 + 23771897 UGCCCG-------CCUUUCCUUUCCACCAACACAAUCCGCAUCAAUAUGUGGUGGAGGGCACACUUAGCCAUUCGCACUUCAUUAGCCCACAUUUCCAAUAUGGUCAUGUGGGUGC---- (((((.-------.........(((((((.((...............))))))))))))))......((.....)).........((((((((..((.....))..))))))))..---- ( -31.86) >DroSec_CAF1 13416 108 + 1 UGCCCG-------CCUUUCA-UUCCACCAACAUAUUCCGCAUCAAUAUGUGGUGGAGUUCACACUUGGCCAUUCGCACUUCAUUAGCCCACUUUUCCAAUAUGUUCAUGUGGGUGC---- (((..(-------((....(-((((((((.((((((.......)))))))))))))))........))).....)))........((((((.................))))))..---- ( -29.13) >DroSim_CAF1 13343 108 + 1 UGCCCG-------CCUUUCA-UUCCACCAACAUAUUCCGCAUCAAUAUGUGGUGGAGUUCACACUUGGCCAUUCGCACUUCAUUAGCCCACAUUUCCAAUAUGGUCAUGUGGGUGC---- (((..(-------((....(-((((((((.((((((.......)))))))))))))))........))).....)))........((((((((..((.....))..))))))))..---- ( -35.80) >DroEre_CAF1 13773 116 + 1 UAGAAGUAAUAGCACUUUCA-UCACACCAACGCAUUUCGAAUCGAUAUGCG---GAGGGCGCACUUAGCCAUUCGCACUUCAUUAGCACACUUUUCCAAUAUGGUUAUGUGGGCGCCCGC ..(((((......)))))..-.........(((((.(((...))).)))))---..((((((.....((.....))............(((....((.....))....))).)))))).. ( -29.80) >DroYak_CAF1 14745 109 + 1 UUCACG-------CCUUUCA-UUACCCCAACAUAUUCGGAAUCAAUAUGUG---GAGGGCACACUUAGCCAUUCGCACUUCAUUAGCACACUUUUCCAAUAUGGUUAUGUGGGCGCCCGC ....((-------(((..((-(.(((.....(((((.((((......((((---((.(((.......))).))))))((.....)).......)))))))))))).))).)))))..... ( -26.00) >consensus UGCCCG_______CCUUUCA_UUCCACCAACAUAUUCCGCAUCAAUAUGUGGUGGAGGGCACACUUAGCCAUUCGCACUUCAUUAGCCCACUUUUCCAAUAUGGUCAUGUGGGUGC____ .....................((((((((.((((((.......))))))))))))))..........((.....)).........((((((....((.....))....))))))...... (-15.18 = -17.82 + 2.64)



| Location | 7,231,795 – 7,231,902 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.97 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -25.25 |
| Energy contribution | -26.45 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7231795 107 + 23771897 AUCAAUAUGUGGUGGAGGGCACACUUAGCCAUUCGCACUUCAUUAGCCCACAUUUCCAAUAUGGUCAUGUGGGUGC-------------CUAGCUUCUGGGCUAAUGGUGUCGUCUUCCG .............(((((((((((...((.....))....(((((((((((((..((.....))..))))))))((-------------((((...))))))))))))))).))))))). ( -41.60) >DroSec_CAF1 13448 107 + 1 AUCAAUAUGUGGUGGAGUUCACACUUGGCCAUUCGCACUUCAUUAGCCCACUUUUCCAAUAUGUUCAUGUGGGUGC-------------CUGGCUUCUGGGCUAAUGGUGUCGUCUUCCG ........(((((.((((....)))).))))).((.((.(((((((((((.....(((...((..(....)..)).-------------.)))....))))))))))).))))....... ( -30.60) >DroSim_CAF1 13375 107 + 1 AUCAAUAUGUGGUGGAGUUCACACUUGGCCAUUCGCACUUCAUUAGCCCACAUUUCCAAUAUGGUCAUGUGGGUGC-------------CUGGCUUCGGGGCUAAUGGUGUCGUCUUCCG ........(((((.((((....)))).))))).((.((.((((((((((((((..((.....))..))))))))((-------------((.(...).)))))))))).))))....... ( -35.90) >DroEre_CAF1 13812 117 + 1 AUCGAUAUGCG---GAGGGCGCACUUAGCCAUUCGCACUUCAUUAGCACACUUUUCCAAUAUGGUUAUGUGGGCGCCCGCUGUUUUCCCCUGGCUUCUGGCUUAAUGGUGUCGUCUUCCG .........((---(((((((((((.(((((...((........(((.(((....((.....))....)))(((....))))))........))...)))))....)))).))))))))) ( -37.19) >DroYak_CAF1 14777 117 + 1 AUCAAUAUGUG---GAGGGCACACUUAGCCAUUCGCACUUCAUUAGCACACUUUUCCAAUAUGGUUAUGUGGGCGCCCGCUGUUUUCCCCCGGCUUCUGGGCUAAUGGUGUCGUCUUCCG ..........(---(((((((((((..(((....((.........)).(((....((.....))....))))))((((((((........))))....))))....))))).))))))). ( -37.30) >consensus AUCAAUAUGUGGUGGAGGGCACACUUAGCCAUUCGCACUUCAUUAGCCCACUUUUCCAAUAUGGUCAUGUGGGUGC_____________CUGGCUUCUGGGCUAAUGGUGUCGUCUUCCG .............(((((((((((((((((...............((((((....((.....))....))))))((................)).....)))))..))))).))))))). (-25.25 = -26.45 + 1.20)



| Location | 7,231,835 – 7,231,938 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.99 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7231835 103 + 23771897 CAUUAGCCCACAUUUCCAAUAUGGUCAUGUGGGUGC-------------CUAGCUUCUGGGCUAAUGGUGUCGUCUUCCGGCAUAAUUGCACUCCAGUGGCAUAGGUUGCCUUCCC---- .....((((((((..((.....))..))))))))((-------------((((((.((((((.(((.((((((.....)))))).)))))...)))).))).))))).........---- ( -38.80) >DroSec_CAF1 13488 97 + 1 CAUUAGCCCACUUUUCCAAUAUGUUCAUGUGGGUGC-------------CUGGCUUCUGGGCUAAUGGUGUCGUCUUCCGGCAUUAUUGGACUCC------CUAGGUUGCCUUCCC---- .....((((((.................))))))..-------------..(((.((((((((((((((((((.....))))))))))))....)------)))))..))).....---- ( -33.03) >DroSim_CAF1 13415 97 + 1 CAUUAGCCCACAUUUCCAAUAUGGUCAUGUGGGUGC-------------CUGGCUUCGGGGCUAAUGGUGUCGUCUUCCGGCAUAAUUGCACUCC------AUAGGUUGCCUUCCC---- .....((((((((..((.....))..))))))))((-------------(((.....(((((.(((.((((((.....)))))).))))).))).------.))))).........---- ( -31.30) >DroEre_CAF1 13849 111 + 1 CAUUAGCACACUUUUCCAAUAUGGUUAUGUGGGCGCCCGCUGUUUUCCCCUGGCUUCUGGCUUAAUGGUGUCGUCUUCCGGCAUAAUUGCACUCCAGUGGAAUAG-----CUACUC---- .....((.(((....((.....))....))).))....(((((..(((.((((......((......((((((.....))))))....))...)))).)))))))-----).....---- ( -29.60) >DroYak_CAF1 14814 115 + 1 CAUUAGCACACUUUUCCAAUAUGGUUAUGUGGGCGCCCGCUGUUUUCCCCCGGCUUCUGGGCUAAUGGUGUCGUCUUCCGGCAUAAUUGCACUCCAGUGGAAUAG-----CGUCUCUCUA ....((.((.((.(((((...(((....(((...((((((((........))))....))))((((.((((((.....)))))).))))))).))).))))).))-----.))))..... ( -32.10) >consensus CAUUAGCCCACUUUUCCAAUAUGGUCAUGUGGGUGC_____________CUGGCUUCUGGGCUAAUGGUGUCGUCUUCCGGCAUAAUUGCACUCCAGUGG_AUAGGUUGCCUUCCC____ (((((((((.....((((.(((....))))))).((................))....)))))))))((((((.....)))))).................................... (-18.55 = -18.99 + 0.44)


| Location | 7,231,902 – 7,232,011 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -16.50 |
| Energy contribution | -17.66 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7231902 109 - 23771897 UGUUACCAAUC-GAACGACACCCUUGUUUGCACUCAAGCCAAAGUGAAAAUCAAUUUACUUAUGCCAUAUGGCA----------GGGAAGGCAACCUAUGCCACUGGAGUGCAAUUAUGC ...........-((((((.....))))))((((((.((((.(((((((......))))))).((((....))))----------))...((((.....)))).)).))))))........ ( -30.30) >DroSec_CAF1 13555 103 - 1 UGUUGUCAAUC-GAACGACACCCUUGUUUGCACUCAAGCCAAAGUGAAUAUCAAUUUACUUACGCCAUAUGGCA----------GGGAAGGCAACCUAG------GGAGUCCAAUAAUGC (((((((....-))..(((.((((.(.((((.(((......((((((((....))))))))..(((....))).----------)))...)))).).))------)).)))))))).... ( -28.10) >DroSim_CAF1 13482 103 - 1 UGUUGCCAAUC-GAACGACACCCUUGUUUGCACUCAAGCCAAAGUGAAAAUCAAUUUACUUAUGCCAUAUGGCA----------GGGAAGGCAACCUAU------GGAGUGCAAUUAUGC (((((......-...))))).......((((((((..(((.(((((((......))))))).((((....))))----------.....)))..(....------)))))))))...... ( -26.60) >DroEre_CAF1 13929 101 - 1 C---UGCAAUC-GAACGACACCCUGGUCUGCACUCAAGCCAAAGUGAAAAUCAAUUUACCCACACCAUAUGGCG----------GAGUAG-----CUAUUCCACUGGAGUGCAAUUAUGC .---.(((...-....(((......)))(((((((..((((..(((................)))....))))(----------((((..-----..)))))....)))))))....))) ( -25.39) >DroYak_CAF1 14894 112 - 1 C---UCCAAUCCAAACGACACCCUAGUCUGCACUCGAGCCAAAGUGAAAAUCAAUUUACCCAUACCAUAUGGCAGCUAUAUAGAGAGACG-----CUAUUCCACUGGAGUGCAAUUAUGC .---............(((......)))(((((((.((.....(((((......))))).........(((((..((........))..)-----))))....)).)))))))....... ( -21.60) >consensus UGUUGCCAAUC_GAACGACACCCUUGUUUGCACUCAAGCCAAAGUGAAAAUCAAUUUACUUAUGCCAUAUGGCA__________GGGAAGGCAACCUAU_CCACUGGAGUGCAAUUAUGC ................(((......)))(((((((..(((((((((((......)))))))........))))...........((........))..........)))))))....... (-16.50 = -17.66 + 1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:43 2006