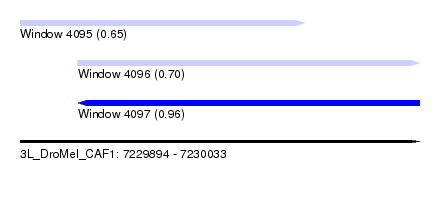
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,229,894 – 7,230,033 |
| Length | 139 |
| Max. P | 0.956856 |

| Location | 7,229,894 – 7,229,993 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -18.54 |
| Energy contribution | -18.23 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7229894 99 + 23771897 ACUUAUAUA--UAACCGUGUA---------------CGUAUGCAUCUAUAUAAUGCGUAGUCUAGUCGCCGAUCUUAGACACUAAGUAGCCAUGUCUGCUGCGCGUUAAUCGCUUU .......((--((...(((((---------------....))))).))))(((((((((((...((.((....(((((...)))))..)).))....)))))))))))........ ( -20.10) >DroSec_CAF1 11609 101 + 1 AACUAUAUAGGUAUACAAGUA---------------GGUAUGUAUGUAUAUAAUGCGUAGUCUAGUCGCUGAUCUUAGACACUAAGUAGCCAUGUCUGCUGCGCGUUAAUCGCUUU ...((((((..(((((.....---------------.)))))..))))))(((((((((((...((.((((..(((((...))))))))).))....)))))))))))........ ( -27.70) >DroSim_CAF1 11526 91 + 1 AACUAUAUA--UAACCAAGUA---------------GGU--------AUAUAAUGCGUAGUCUAGUCGCCGAUCUUAGACACUAAGUAGCCAUGUCUGCUGCGCGUUAAUCGCUUU .........--..(((.....---------------)))--------...(((((((((((...((.((....(((((...)))))..)).))....)))))))))))........ ( -20.50) >DroEre_CAF1 11830 106 + 1 ACCUAUAUA--UACGCAUAUAUAUAUCCGAUAUAUAC--------GUAUACGAUGCGUGGUCUCGUCGCCGAUCUUAGACACUAAGUAGCCAUGUCUGCCGCGCGUUAAUCGCUUU .......((--((((.((((((.......)))))).)--------))))).((((((((((..(((.((....(((((...)))))..)).)))...))))))))))......... ( -29.40) >consensus AACUAUAUA__UAACCAAGUA_______________CGU______GUAUAUAAUGCGUAGUCUAGUCGCCGAUCUUAGACACUAAGUAGCCAUGUCUGCUGCGCGUUAAUCGCUUU ..................................................(((((((((((...((.((....(((((...)))))..)).))....)))))))))))........ (-18.54 = -18.23 + -0.31)

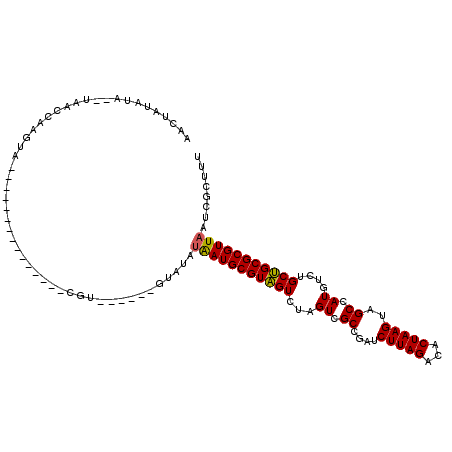

| Location | 7,229,914 – 7,230,033 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -31.54 |
| Energy contribution | -31.35 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7229914 119 + 23771897 GUAUGCAUCUAUAUAAUGCGUAGUCUAGUCGCCGAUCUUAGACACUAAGUAGCCAUGUCUGCUGCGCGUUAAUCGCUUUUAAUUGCUAAAUUACGCGCUGCAUGGCCGACAUUGCGACG .(((((((.......))))))).....(((((....(((((...)))))..(((((((.....((((((.....((........))......)))))).))))))).......))))). ( -37.40) >DroSec_CAF1 11631 119 + 1 GUAUGUAUGUAUAUAAUGCGUAGUCUAGUCGCUGAUCUUAGACACUAAGUAGCCAUGUCUGCUGCGCGUUAAUCGCUUUUAAUUGCUAAAUUACGCGCUGCAUGGCCGACAUUGCGACG .(((((((.......))))))).....(((((....(((((...)))))..(((((((.....((((((.....((........))......)))))).))))))).......))))). ( -37.00) >DroSim_CAF1 11546 111 + 1 GU--------AUAUAAUGCGUAGUCUAGUCGCCGAUCUUAGACACUAAGUAGCCAUGUCUGCUGCGCGUUAAUCGCUUUUAAUUGCUAAAUUACGCGCUGCAUGGCCGACAUUGCGACG ..--------........((((((...((((.....(((((...)))))..(((((((.....((((((.....((........))......)))))).)))))))))))))))))... ( -32.00) >DroEre_CAF1 11865 111 + 1 --------GUAUACGAUGCGUGGUCUCGUCGCCGAUCUUAGACACUAAGUAGCCAUGUCUGCCGCGCGUUAAUCGCUUUUAAUUGCUAAAUUACGCGCUGCAUGGUCGACAUUGCGACG --------...((((...))))....(((((((((((..(((((...........)))))((.((((((.....((........))......)))))).))..))))).....)))))) ( -31.60) >consensus GU______GUAUAUAAUGCGUAGUCUAGUCGCCGAUCUUAGACACUAAGUAGCCAUGUCUGCUGCGCGUUAAUCGCUUUUAAUUGCUAAAUUACGCGCUGCAUGGCCGACAUUGCGACG ...........................(((((....(((((...)))))..(((((((.....((((((.....((........))......)))))).))))))).......))))). (-31.54 = -31.35 + -0.19)

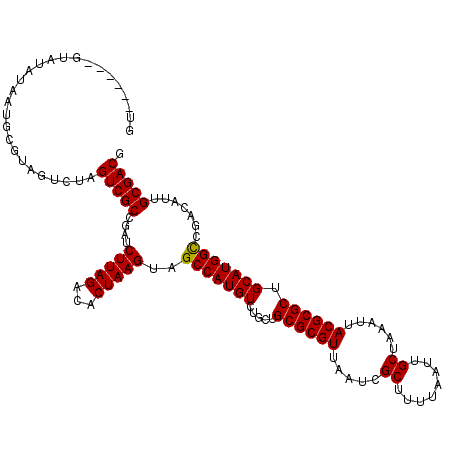
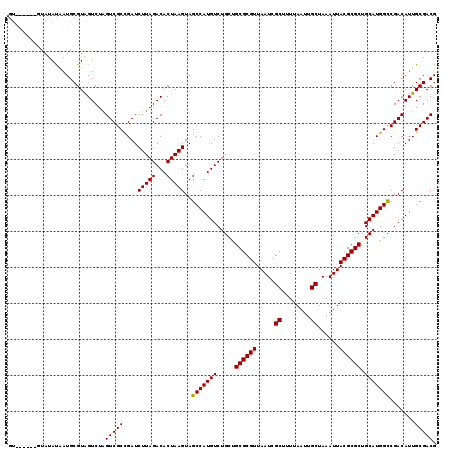
| Location | 7,229,914 – 7,230,033 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -31.83 |
| Energy contribution | -32.08 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7229914 119 - 23771897 CGUCGCAAUGUCGGCCAUGCAGCGCGUAAUUUAGCAAUUAAAAGCGAUUAACGCGCAGCAGACAUGGCUACUUAGUGUCUAAGAUCGGCGACUAGACUACGCAUUAUAUAGAUGCAUAC .(((((......((((((((.((((((((((..((........)))))).)))))).)))....))))).(((((...)))))....)))))........(((((.....))))).... ( -37.90) >DroSec_CAF1 11631 119 - 1 CGUCGCAAUGUCGGCCAUGCAGCGCGUAAUUUAGCAAUUAAAAGCGAUUAACGCGCAGCAGACAUGGCUACUUAGUGUCUAAGAUCAGCGACUAGACUACGCAUUAUAUACAUACAUAC .(((((......((((((((.((((((((((..((........)))))).)))))).)))....))))).(((((...)))))....)))))........................... ( -33.30) >DroSim_CAF1 11546 111 - 1 CGUCGCAAUGUCGGCCAUGCAGCGCGUAAUUUAGCAAUUAAAAGCGAUUAACGCGCAGCAGACAUGGCUACUUAGUGUCUAAGAUCGGCGACUAGACUACGCAUUAUAU--------AC .(((((......((((((((.((((((((((..((........)))))).)))))).)))....))))).(((((...)))))....))))).................--------.. ( -33.10) >DroEre_CAF1 11865 111 - 1 CGUCGCAAUGUCGACCAUGCAGCGCGUAAUUUAGCAAUUAAAAGCGAUUAACGCGCGGCAGACAUGGCUACUUAGUGUCUAAGAUCGGCGACGAGACCACGCAUCGUAUAC-------- ((((((...(((......((.((((((((((..((........)))))).)))))).))((((((.........))))))..)))..))))))..................-------- ( -33.50) >consensus CGUCGCAAUGUCGGCCAUGCAGCGCGUAAUUUAGCAAUUAAAAGCGAUUAACGCGCAGCAGACAUGGCUACUUAGUGUCUAAGAUCGGCGACUAGACUACGCAUUAUAUAC______AC .(((((......((((((((.((((((((((..((........)))))).)))))).)))....))))).(((((...)))))....)))))........................... (-31.83 = -32.08 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:34 2006