| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,228,948 – 7,229,039 |
| Length | 91 |
| Max. P | 0.964731 |

| Location | 7,228,948 – 7,229,039 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -19.28 |
| Energy contribution | -19.16 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7228948 91 + 23771897 UGAGAAACCAG------GCGAGGCGCCACCGCCA-AGUGAAUGACCACAAAAAUGCACUCCGCUGCA------AGAGUUGUGGUC----------------AAGUGUCGUAAAGGUUGAA .....((((..------((((((((....)))).-......(((((((((...((((......))))------....))))))))----------------)....))))...))))... ( -31.60) >DroSec_CAF1 10647 91 + 1 UGAGAAACCAG------GCGAGGCGCCAUCGCCA-AGUGAAUGACCACAAAAAUGCACUCCACUGCA------AGAGUUGUGGUC----------------AAGUGUCGUAAAGGUUGAA .....((((..------((((((((....)))).-......(((((((((...((((......))))------....))))))))----------------)....))))...))))... ( -31.40) >DroSim_CAF1 10565 92 + 1 UGAGAAACCAG------GCGAGGCGCCAUCGCCAAAGUGAAUGACCACAAAAAUGCACUCCACUGCA------AGAGUUGUGGUC----------------AAGUGUCGUAAAGGUUGAA .....((((..------((((.((....((((....)))).(((((((((...((((......))))------....))))))))----------------).)).))))...))))... ( -32.10) >DroEre_CAF1 10790 94 + 1 UGAGAAAACUGUGAAA-CU--GGCGCCGCCGCCA-AGGGAGUGACCACAAAAAUGCACUCCACUGCA------ACCGUUGUGGUC----------------AAGUGUCGUAAAGGUUGAA ......((((.(((.(-((--((((....)))).-......(((((((((...((((......))))------....))))))))----------------)))).)))....))))... ( -27.50) >DroYak_CAF1 11583 119 + 1 UGAGAAAAAAAAAAAAGCUAAGGCGCCGUCGCCA-AGGGAAUGACCACAAAAAUGCACUCCACUGCACUGACGACCGUUGUGGUCAAGUGGGCAAGUGGUCAAGUGUCGUUAAGGUUGAA ...............((((..((((....)))).-((.((.(((((((.....(((...(((((....((((.((....)).)))))))))))).)))))))....)).))..))))... ( -30.50) >consensus UGAGAAACCAG______GCGAGGCGCCAUCGCCA_AGUGAAUGACCACAAAAAUGCACUCCACUGCA______AGAGUUGUGGUC________________AAGUGUCGUAAAGGUUGAA .....((((........((..((((....))))...))....((((((((...((((......))))..........))))))))............................))))... (-19.28 = -19.16 + -0.12)

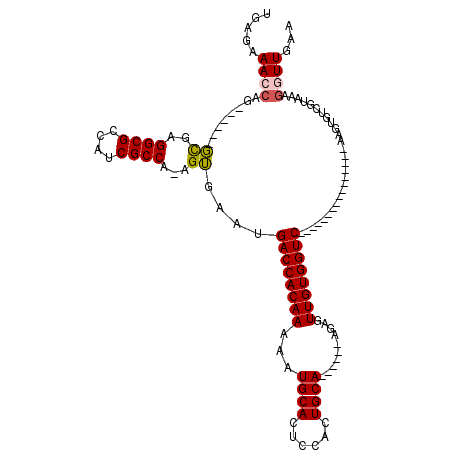

| Location | 7,228,948 – 7,229,039 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.73 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7228948 91 - 23771897 UUCAACCUUUACGACACUU----------------GACCACAACUCU------UGCAGCGGAGUGCAUUUUUGUGGUCAUUCACU-UGGCGGUGGCGCCUCGC------CUGGUUUCUCA ...((((....(((....(----------------((((((((....------((((......))))...)))))))))......-.((((....))))))).------..))))..... ( -27.30) >DroSec_CAF1 10647 91 - 1 UUCAACCUUUACGACACUU----------------GACCACAACUCU------UGCAGUGGAGUGCAUUUUUGUGGUCAUUCACU-UGGCGAUGGCGCCUCGC------CUGGUUUCUCA ...((((.....((....(----------------((((((((....------((((......))))...))))))))).))...-.(((((.(....)))))------).))))..... ( -27.90) >DroSim_CAF1 10565 92 - 1 UUCAACCUUUACGACACUU----------------GACCACAACUCU------UGCAGUGGAGUGCAUUUUUGUGGUCAUUCACUUUGGCGAUGGCGCCUCGC------CUGGUUUCUCA ...((((.....((....(----------------((((((((....------((((......))))...))))))))).)).....(((((.(....)))))------).))))..... ( -27.90) >DroEre_CAF1 10790 94 - 1 UUCAACCUUUACGACACUU----------------GACCACAACGGU------UGCAGUGGAGUGCAUUUUUGUGGUCACUCCCU-UGGCGGCGGCGCC--AG-UUUCACAGUUUUCUCA ............((.((.(----------------((((((((..((------.(((......)))))..)))))))))......-(((((....))))--))-).))............ ( -26.00) >DroYak_CAF1 11583 119 - 1 UUCAACCUUAACGACACUUGACCACUUGCCCACUUGACCACAACGGUCGUCAGUGCAGUGGAGUGCAUUUUUGUGGUCAUUCCCU-UGGCGACGGCGCCUUAGCUUUUUUUUUUUUCUCA ..................(((((((.((((((((.((((.....))))((....)))))))...))).....)))))))......-.((((....))))..................... ( -29.00) >consensus UUCAACCUUUACGACACUU________________GACCACAACUCU______UGCAGUGGAGUGCAUUUUUGUGGUCAUUCACU_UGGCGAUGGCGCCUCGC______CUGGUUUCUCA ...................................((((((((..........((((......))))...)))))))).........((((....))))..................... (-18.48 = -18.48 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:30 2006