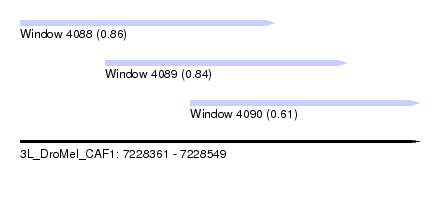
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,228,361 – 7,228,549 |
| Length | 188 |
| Max. P | 0.859690 |

| Location | 7,228,361 – 7,228,481 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -20.68 |
| Energy contribution | -20.56 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7228361 120 + 23771897 UAUUUGUUCCCGCAUUUACCAUUUACCAUUUACCUUGGCACUCCGAGAGGCCUUAACAUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGAAAGCCUCGAGAACUCACUCGAGAGCAC ..((((((...((....................(((((....))))).(((.(((.....))).))).........))...))))))........((((((((......)))))).)).. ( -24.10) >DroSec_CAF1 10072 113 + 1 UGUUUGUUCCCGCAUUUGC-------CAUUUACCUUGGCGCACCGAGAGGCCUUAACAUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGAAAGUCUCGAGAACUCACUCGAGAGCAU .(((((((...((....((-------((.......)))).........(((.(((.....))).))).........))...)))))))........(((((((......))))))).... ( -26.20) >DroSim_CAF1 9982 120 + 1 UGUUUGUUCCCCCAUUUACCAUUUACCAUUUACCUUGGCGCACCGAGAGGCCUUAACAUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGAAAGCCUCGAGAACUCACUCGAGAGCAU (((((((..........................(((((....))))).(((.(((.....))).)))..)))))))...................((((((((......)))))).)).. ( -25.40) >DroEre_CAF1 10089 113 + 1 UGCUUGUUCCCGCAUUUAC-------CAUUCACCUUGCCGCACCGCGUGGCCUUAACAUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGAAAGCCUCGAGAACUCGCUCGAGAGCAU (((........))).....-------..(((((...((......))(((((.(((.....))).)))))....................))))).((((((((......)))))).)).. ( -27.60) >DroYak_CAF1 11035 120 + 1 UGCUUGUUCCCGCAUUUACCAUCUACCAUUUACCUUGCCACAUCGAGUGGCCUUAACAUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGAAAGCCUCGAGAACUCACUCGAGAGCAU ((((.......((.(((((.................(((((.....)))))........((((.((..........)).))))......))))).))((((((......)))))))))). ( -24.80) >consensus UGUUUGUUCCCGCAUUUACCAU_UACCAUUUACCUUGGCGCACCGAGAGGCCUUAACAUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGAAAGCCUCGAGAACUCACUCGAGAGCAU ............................(((((((((......)))).(((.(((.....))).)))......................))))).((((((((......)))))).)).. (-20.68 = -20.56 + -0.12)

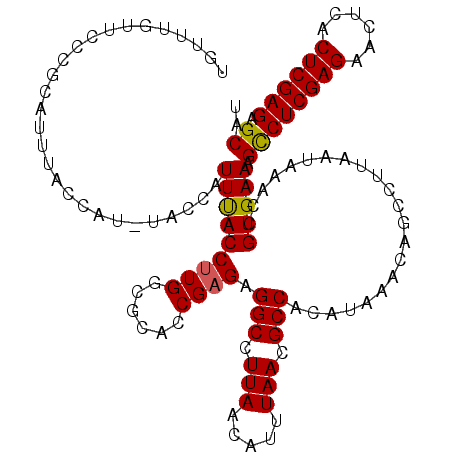
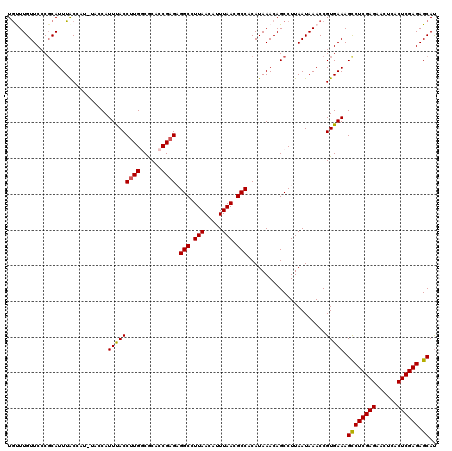
| Location | 7,228,401 – 7,228,515 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.29 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -18.93 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7228401 114 + 23771897 CUCCGAGAGGCCUUAACAUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGAAAGCCUCGAGAACUCACUCGAGAGCACCGUUAGUUUCAUAUAUAUAUUUAUAUAUGUACUA------ ...((...(((.(((.....))).)))............................((((((((......)))))).))..)).((((..((((((((....)))))))).))))------ ( -24.50) >DroEre_CAF1 10122 110 + 1 CACCGCGUGGCCUUAACAUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGAAAGCCUCGAGAACUCGCUCGAGAGCAUCGUUAGUUUCAUGCACAAAGAUAU----AUUCUA------ ....(((((((.(((.....))).)))))....................((((((((((((((......)))))).))..(....)))))))))....(((...----..))).------ ( -25.30) >DroYak_CAF1 11075 116 + 1 CAUCGAGUGGCCUUAACAUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGAAAGCCUCGAGAACUCACUCGAGAGCAUCGUUAGUUGCAAGCACAAAAAUAU----AUAAUAUAAUAU ......(((((.(((.....))).)))))....................(((...((((((((......)))))).))...((.....))...)))........----............ ( -23.30) >consensus CACCGAGUGGCCUUAACAUUUAACGCCACAUAAACAGCCUUAAUAAACCGUGAAAGCCUCGAGAACUCACUCGAGAGCAUCGUUAGUUUCAUGCACAAAAAUAU____AUACUA______ ......(((((.(((.....))).)))))..........................((((((((......)))))).)).......................................... (-18.93 = -19.27 + 0.33)

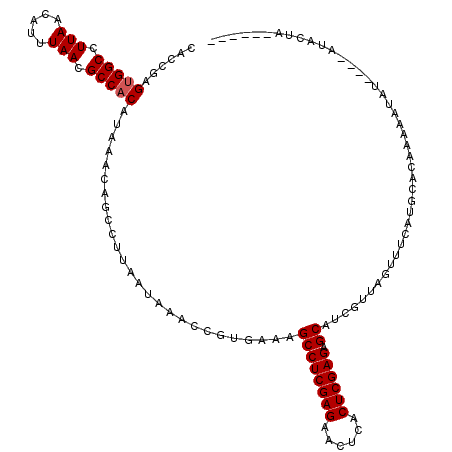
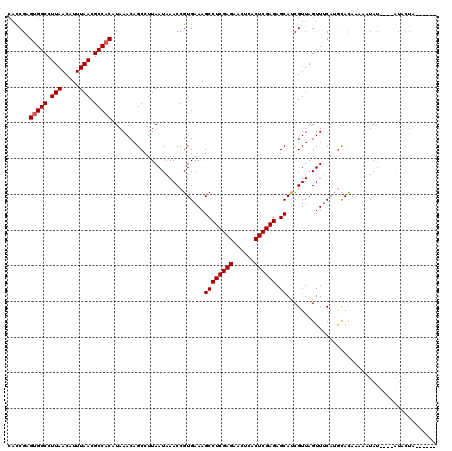
| Location | 7,228,441 – 7,228,549 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.51 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.00 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
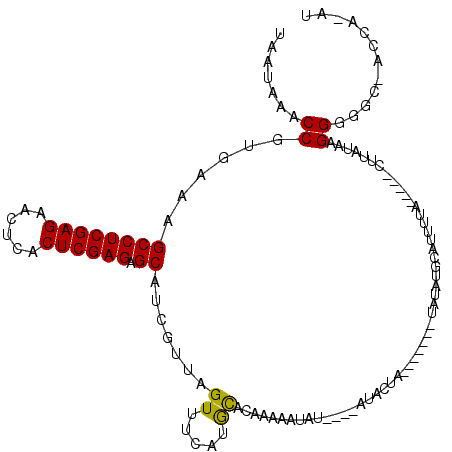
>3L_DroMel_CAF1 7228441 108 + 23771897 UAAUAAACCGUGAAAGCCUCGAGAACUCACUCGAGAGCACCGUUAGUUUCAUAUAUAUAUUUAUAUAUGUACUA-------UAUAUGCAUGUUA-----CUUAUAAGGAGGCAACCAUAU .........(((...((((((((......)))....(((....((((..((((((((....)))))))).))))-------....)))......-----........)))))...))).. ( -25.30) >DroEre_CAF1 10162 101 + 1 UAAUAAACCGUGAAAGCCUCGAGAACUCGCUCGAGAGCAUCGUUAGUUUCAUGCACAAAGAUAU----AUUCUA-------UAUACCAAUUUUU-----CUUAUAAGGGGGC-ACC--AU .........((((((((((((((......)))))).))..(....)))))))((....(((...----..))).-------....((..((...-----.....))..))))-...--.. ( -17.60) >DroYak_CAF1 11115 115 + 1 UAAUAAACCGUGAAAGCCUCGAGAACUCACUCGAGAGCAUCGUUAGUUGCAAGCACAAAAAUAU----AUAAUAUAAUAUGUAUAUGCAUUUUACAUAUGUUAUAAGGGGUU-ACCACAU .....(((((((...((((((((......)))))).))...((.....))...))).......(----(((((...(((((((.........)))))))))))))...))))-....... ( -24.80) >consensus UAAUAAACCGUGAAAGCCUCGAGAACUCACUCGAGAGCAUCGUUAGUUUCAUGCACAAAAAUAU____AUACUA_______UAUAUGCAUUUUA_____CUUAUAAGGGGGC_ACCA_AU .......((......((((((((......)))))).)).......((.....))....................................................))............ (-13.44 = -13.00 + -0.44)

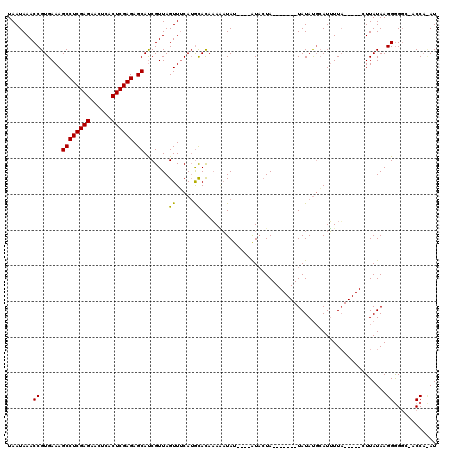
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:28 2006