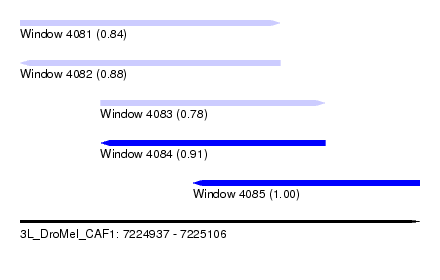
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 7,224,937 – 7,225,106 |
| Length | 169 |
| Max. P | 0.997885 |

| Location | 7,224,937 – 7,225,047 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -21.17 |
| Energy contribution | -22.50 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.42 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7224937 110 + 23771897 AUAAGUGGUUUAUAUGUGGCUAUUCAUAUUUCUUCUCCGCCCAAAGGAUCCCCUUUCAUCAAACCGCAUAUGUAUGUACAUACAUACAUAUGUACAUAUGCAUAACAAUC ....(((((((...((.(((..................)))))((((....)))).....))))))).((((((((((((((......)))))))))..)))))...... ( -26.17) >DroSec_CAF1 6630 102 + 1 AUAAGUGGUUUAUAUGUGGCUAUUCAUAUUUCUUCUCCGCCCAAAGGAUCCCCUUUCAUCAAACCGCAUAUGUAUGUACAUACAU--------ACAUAUGCAUAACAAUC ......(((((...((.(((..................)))))((((....)))).....)))))((((((((((((....))))--------))))))))......... ( -27.07) >DroSim_CAF1 6583 106 + 1 AUAAGUGGUUUAUAUGUGGCUAUUCAUAUUUCUUCUCCGCCCAAAGGAUCCCCUUUCAUCAAACCGCAUAUGUAUGUACAUACAU----AUGUACAUAUGCAUAACAAUC ......(((((...((.(((..................)))))((((....)))).....)))))(((((((((((((......)----))))))))))))......... ( -25.27) >consensus AUAAGUGGUUUAUAUGUGGCUAUUCAUAUUUCUUCUCCGCCCAAAGGAUCCCCUUUCAUCAAACCGCAUAUGUAUGUACAUACAU____AUGUACAUAUGCAUAACAAUC ....(((((((...((.(((..................)))))((((....)))).....))))))).(((((((((((((........))))))))..)))))...... (-21.17 = -22.50 + 1.33)

| Location | 7,224,937 – 7,225,047 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -28.94 |
| Consensus MFE | -24.47 |
| Energy contribution | -24.47 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7224937 110 - 23771897 GAUUGUUAUGCAUAUGUACAUAUGUAUGUAUGUACAUACAUAUGCGGUUUGAUGAAAGGGGAUCCUUUGGGCGGAGAAGAAAUAUGAAUAGCCACAUAUAAACCACUUAU .........(((((((((....(((((....))))))))))))))((((((((((((((....))))).(((..................))).))).))))))...... ( -28.67) >DroSec_CAF1 6630 102 - 1 GAUUGUUAUGCAUAUGU--------AUGUAUGUACAUACAUAUGCGGUUUGAUGAAAGGGGAUCCUUUGGGCGGAGAAGAAAUAUGAAUAGCCACAUAUAAACCACUUAU .........((((((((--------((((....))))))))))))((((((((((((((....))))).(((..................))).))).))))))...... ( -31.47) >DroSim_CAF1 6583 106 - 1 GAUUGUUAUGCAUAUGUACAU----AUGUAUGUACAUACAUAUGCGGUUUGAUGAAAGGGGAUCCUUUGGGCGGAGAAGAAAUAUGAAUAGCCACAUAUAAACCACUUAU .........(((((((((..(----((....)))..)))))))))((((((((((((((....))))).(((..................))).))).))))))...... ( -26.67) >consensus GAUUGUUAUGCAUAUGUACAU____AUGUAUGUACAUACAUAUGCGGUUUGAUGAAAGGGGAUCCUUUGGGCGGAGAAGAAAUAUGAAUAGCCACAUAUAAACCACUUAU .........((((((((........((((....))))))))))))((((((((((((((....))))).(((..................))).))).))))))...... (-24.47 = -24.47 + 0.00)

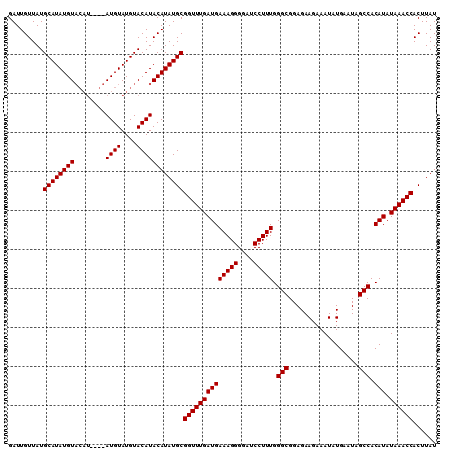
| Location | 7,224,971 – 7,225,066 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -17.37 |
| Energy contribution | -18.70 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7224971 95 + 23771897 CUCCGCCCAAAGGAUCCCCUUUCAUCAAACCGCAUAUGUAUGUACAUACAUACAUAUGUACAUAUGCAUAACAAUCAGUAUCGCUGGCUCUCGCG ...(((..(((((....))))).........((((((((((((....))))))))))))...............((((.....)))).....))) ( -22.30) >DroSec_CAF1 6664 87 + 1 CUCCGCCCAAAGGAUCCCCUUUCAUCAAACCGCAUAUGUAUGUACAUACAU--------ACAUAUGCAUAACAAUCAGUAUCGCUGGCUCUCGCG ...(((..(((((....))))).........((((((((((((....))))--------)))))))).......((((.....)))).....))) ( -24.70) >DroSim_CAF1 6617 91 + 1 CUCCGCCCAAAGGAUCCCCUUUCAUCAAACCGCAUAUGUAUGUACAUACAU----AUGUACAUAUGCAUAACAAUCCGUAUCGCUGGCUCUCGCG ....(((.(((((....))))).........(((((((((((((......)----))))))))))))..................)))....... ( -22.50) >consensus CUCCGCCCAAAGGAUCCCCUUUCAUCAAACCGCAUAUGUAUGUACAUACAU____AUGUACAUAUGCAUAACAAUCAGUAUCGCUGGCUCUCGCG ....(((.(((((....))))).........((((((((((((............))))))))))))..................)))....... (-17.37 = -18.70 + 1.33)



| Location | 7,224,971 – 7,225,066 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -25.60 |
| Energy contribution | -25.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7224971 95 - 23771897 CGCGAGAGCCAGCGAUACUGAUUGUUAUGCAUAUGUACAUAUGUAUGUAUGUACAUACAUAUGCGGUUUGAUGAAAGGGGAUCCUUUGGGCGGAG (((..(((((((((((....))))))..(((((((((....(((((....)))))))))))))))))))..(.(((((....))))).))))... ( -29.80) >DroSec_CAF1 6664 87 - 1 CGCGAGAGCCAGCGAUACUGAUUGUUAUGCAUAUGU--------AUGUAUGUACAUACAUAUGCGGUUUGAUGAAAGGGGAUCCUUUGGGCGGAG (((..(((((((((((....))))))..((((((((--------((((....)))))))))))))))))..(.(((((....))))).))))... ( -32.60) >DroSim_CAF1 6617 91 - 1 CGCGAGAGCCAGCGAUACGGAUUGUUAUGCAUAUGUACAU----AUGUAUGUACAUACAUAUGCGGUUUGAUGAAAGGGGAUCCUUUGGGCGGAG (((..(((((((((((....))))))..(((((((((..(----((....)))..))))))))))))))..(.(((((....))))).))))... ( -27.80) >consensus CGCGAGAGCCAGCGAUACUGAUUGUUAUGCAUAUGUACAU____AUGUAUGUACAUACAUAUGCGGUUUGAUGAAAGGGGAUCCUUUGGGCGGAG (((..(((((((((((....))))))..((((((((........((((....)))))))))))))))))..(.(((((....))))).))))... (-25.60 = -25.60 + -0.00)

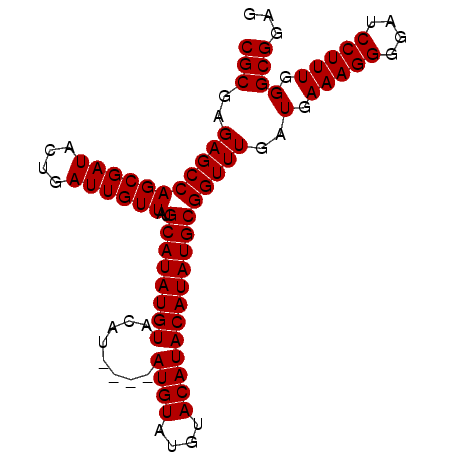

| Location | 7,225,010 – 7,225,106 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -21.67 |
| Energy contribution | -22.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7225010 96 - 23771897 UCGCUAUUCGGUUAGCAAGCUGAGUGGCAAAUACUCGGAUCGCGAGAGCCAGCGAUACUGAUUGUUAUGCAUAUGUACAUAUGUAUGUAUGUACAU ..(((((((((((....))))))))))).((((.(((((((((........))))).)))).)))).(((((((((((....)))))))))))... ( -37.40) >DroSec_CAF1 6703 88 - 1 UCGCUAUUCGGUUAGCAAGCUGAGUGGCAAAUACUCGGAUCGCGAGAGCCAGCGAUACUGAUUGUUAUGCAUAUGU--------AUGUAUGUACAU ..(((((((((((....))))))))))).((((.(((((((((........))))).)))).)))).(((((((..--------..)))))))... ( -31.70) >DroSim_CAF1 6656 92 - 1 UCGCUAUUCGGUUAGCAAGCUGAGUGGCAAAUACUCCGAUCGCGAGAGCCAGCGAUACGGAUUGUUAUGCAUAUGUACAU----AUGUAUGUACAU ..(((((((((((....)))))))))))......(((((((((........))))).)))).((((((((((((....))----))))))).))). ( -36.10) >DroAna_CAF1 6539 86 - 1 UCGGUAUUCGGUCAGCAAGCUGAGUGGAAAAUAUUCGGAUCGGGAGAGCCAGCGAUACUAAUCCAGAUGCGAAGACCCAU----CUGCAA------ ..(((.((((.((((....)))).)))).....((((.(((.(((................))).))).)))).)))...----......------ ( -21.39) >consensus UCGCUAUUCGGUUAGCAAGCUGAGUGGCAAAUACUCGGAUCGCGAGAGCCAGCGAUACUGAUUGUUAUGCAUAUGUACAU____AUGUAUGUACAU ..((((((((((......))))))))))......(((((((((........))))).))))....(((((((............)))))))..... (-21.67 = -22.68 + 1.00)

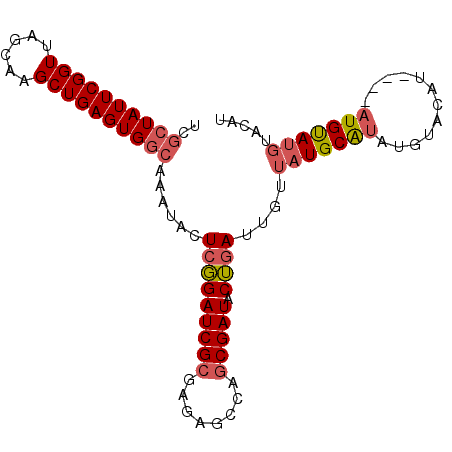
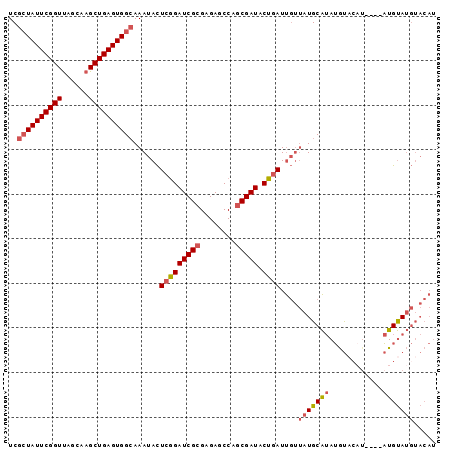
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:23 2006