| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
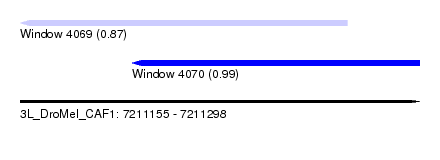
| Location | 7,211,155 – 7,211,298 |
| Length | 143 |
| Max. P | 0.994964 |

| Location | 7,211,155 – 7,211,272 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.32 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -17.86 |
| Energy contribution | -19.94 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
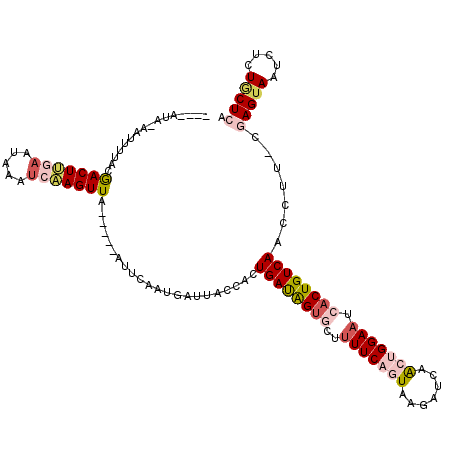
>3L_DroMel_CAF1 7211155 117 - 23771897 -AUUCAAUGAUUACCACUGAUAGUGCUUUUCAGUAAGAUCAACUGGAAUCACUGUCAACUUU-CGAGUA-UAUCUGCUCACAUGAACAAUCUAGUUACUCCGUUGAAUUUUUAACACUAA -((((((((........((((((((..(((((((.......))))))).)))))))).....-.(((((-....))))).....................))))))))............ ( -27.60) >DroSec_CAF1 4286 118 - 1 -AUUCAAUGAUUACCGCUGAUAGUGCUUUUCAGUAAGUUCAACUGGAAUCACUGUCAACCUU-CGAGUAAUCUCUGCUCACAUGAACAAUCUAGGUACUCUGCUGAAUUUUUAACACUAA -(((((..((.((((..((((((((..(((((((.......))))))).)))))))).....-.(((((.....)))))..............)))).))...)))))............ ( -29.80) >DroSim_CAF1 4312 118 - 1 -AUUCAAUGAUUACCACUGACAGUGCUUUUCAGUAAGAUCAACUGGAAUCACUGUCAACCUU-CCAGUAAUCUCUGCUCACAUGAACAAUCUAGGUACUCUGUUGAAUUUUUAACACUAA -(((((((((.((((..((((((((..(((((((.......))))))).)))))))).....-.(((......))).................)))).)).)))))))............ ( -30.00) >DroEre_CAF1 848 118 - 1 AAUUUCAUGAUUACCACUGACGGUACUUUUCAGUAUGAUCAAAAGGACUCACUGUCAACGCU-CCAG-AAUCUCUGCUCACAUGAACAAUCUAGGUACUGCGAUGCAUUUUUAACACUAA .......((((((..(((((.........))))).))))))....(((.....)))...(((-((((-.((((.((.((....)).))....)))).))).)).)).............. ( -19.20) >DroYak_CAF1 213 120 - 1 AAUUUAUUGACUACCACUGAUAGCACUUUUCAGUAAGAUCAGAAGGAAUCACUGUCAACCCCUGGAGUAAUCUCUACUCACAUGAACAAUCUAGGUACUCCGAUGCAUUUUUAACACUAA ......(((((.....(((((...(((....)))...))))).((......)))))))....(((((((.(((....((....)).......)))))))))).................. ( -17.30) >consensus _AUUCAAUGAUUACCACUGAUAGUGCUUUUCAGUAAGAUCAACUGGAAUCACUGUCAACCUU_CGAGUAAUCUCUGCUCACAUGAACAAUCUAGGUACUCCGAUGAAUUUUUAACACUAA .((((((((..((((..((((((((..(((((((.......))))))).)))))))).......(((((.....)))))..............))))...))))))))............ (-17.86 = -19.94 + 2.08)



| Location | 7,211,195 – 7,211,298 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.57 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -16.02 |
| Energy contribution | -17.86 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 7211195 103 - 23771897 ----------UGUUACGACUUGAAUAAAUCAAGUUA-----AUUCAAUGAUUACCACUGAUAGUGCUUUUCAGUAAGAUCAACUGGAAUCACUGUCAACUUU-CGAGUA-UAUCUGCUCA ----------......(((((((.....))))))).-----................((((((((..(((((((.......))))))).)))))))).....-.(((((-....))))). ( -27.20) >DroSec_CAF1 4326 110 - 1 ----AUACAAUUUUACGACUUGAAUAAAUCAAGUUA-----AUUCAAUGAUUACCGCUGAUAGUGCUUUUCAGUAAGUUCAACUGGAAUCACUGUCAACCUU-CGAGUAAUCUCUGCUCA ----............(((((((.....))))))).-----...((..((((((((.((((((((..(((((((.......))))))).)))))))).....-)).))))))..)).... ( -28.80) >DroSim_CAF1 4352 110 - 1 ----AUAUAAUUUUGCGACUUGAAUAAAUCAAGUUA-----AUUCAAUGAUUACCACUGACAGUGCUUUUCAGUAAGAUCAACUGGAAUCACUGUCAACCUU-CCAGUAAUCUCUGCUCA ----..........(((((((((.....))))))).-----.......((((((...((((((((..(((((((.......))))))).)))))))).....-...))))))...))... ( -29.00) >DroEre_CAF1 888 117 - 1 CCAUAUAUAAGUGUACAACUCUAAUAAACCCAGUUA-UUGAAUUUCAUGAUUACCACUGACGGUACUUUUCAGUAUGAUCAAAAGGACUCACUGUCAACGCU-CCAG-AAUCUCUGCUCA .........(((((.......((((((......)))-))).......((((((..(((((.........))))).))))))....(((.....))).)))))-.(((-.....))).... ( -16.10) >DroYak_CAF1 253 112 - 1 --------CAGUUUACAACUGCAAUAAAUCCAGUUAAUUGAAUUUAUUGACUACCACUGAUAGCACUUUUCAGUAAGAUCAGAAGGAAUCACUGUCAACCCCUGGAGUAAUCUCUACUCA --------..(((.(((....((((((((.(((....))).))))))))....((.(((((...(((....)))...)))))..))......))).))).....(((((.....))))). ( -19.80) >consensus ____AUA_AAUUUUACGACUUGAAUAAAUCAAGUUA_____AUUCAAUGAUUACCACUGAUAGUGCUUUUCAGUAAGAUCAACUGGAAUCACUGUCAACCUU_CGAGUAAUCUCUGCUCA ................(((((((.....)))))))......................((((((((..(((((((.......))))))).)))))))).......(((((.....))))). (-16.02 = -17.86 + 1.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:10 2006